Gene transcriptions/Start sites
< Gene transcriptions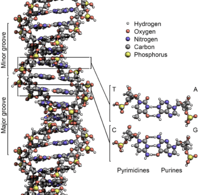
The transcription start site "is the location where transcription starts at the 5'-end of a gene sequence.[1]"[2]
Each human gene is made up of deoxyribonucleic acid (DNA) in a double helix. Along each helix which is composed of a phosphate-deoxyribose polymer are nitrogenous bases. These bases are linked across the helices by hydrogen bonds, one bond per nitrogenous base pair (bp). Each nitrogenous base is part of a nucleotide (nt).
Notations
Notation: let the symbol bp indicate a nitrogenous nucleobase pair as linked by a hydrogen bond between the two antiparallel helices that compose DNA.
Notation: let the symbol nt indicate a nucleotide.
Notation: for the coding of individual nucleobases along a DNA strand, the following letters are standardized by convention:
- adenine - A,
- cytosine - C,
- guanine - G, and
- thymine - T.
Notation: let the subscript (+1) indicate the specific nucleobase along the template strand that is a transcription start site. For example, A+1.
Nitrogenous bases
Nitrogenous "bases are typically classified as the derivatives of two parent compounds, pyrimidine and purine.[3]"[4]
"Three nucleobases found in nucleic acids, cytosine (C), thymine (T), and uracil (U), are pyrimidine derivatives".[5] Of these, cytosine (C) and thymine (T) occur in DNA.
"Two of the four bases in nucleic acids, adenine (2) and guanine (3), are purines."[6]
The four nitrogenous bases that occur in DNA linking between the two phosphate-deoxyribose polymer strands are adenine (A), cytosine (C), guanine (G), and thymine (T).
Gene expressions
Although it is harder to regulate the transcription of genes with multiple transcription start sites, "variations in the expression of a constitutive gene would be minimized by the use of multiple start sites."[7]
Earlier "studies led to the design of a super core promoter (SCP) that contains a TATA, Inr, MTE, and DPE in a single promoter (Juven-Gershon et al., 2006b). The SCP is the strongest core promoter observed in vitro and in cultured cells and yields high levels of transcription in conjunction with transcriptional enhancers. These findings indicate that gene expression levels can be modulated via the core promoter."[7]
Gene transcriptions
"From a teleological standpoint, this arrangement [of focused promoters] is consistent with the notion that it would be easier to regulate the transcription of a gene with a single transcription start site than one with multiple start sites."[7]
"Bioinformatics programs usually allow for alternate start codons when searching for protein coding genes."[8]
RNA polymerase II holoenzyme complex may also have to search for one or more transcription start sites.
"RNA polymerase II has been redirected to alternative start sites by reducing ATP concentrations within a nuclear extract, by altering the spacing between the TATA and Inr in a promoter containing both elements, and by dinucleotide initiation strategies".[9]
The core promoter includes the transcription start site(s) (TSS).
RNA polymerase II holoenzyme complexes
"RNA polymerase II ... is recruited to the promoters of protein-coding genes in living cells.[10]"[11] Or, transcription factories are present and the euchromatin is brought within the nearest transcription factory and A1BG messenger RNA (mRNA) is transcribed.
For those circumstances in which the holoenzyme is built onto the euchromatin, it is necessary to consider the holoenzyme components and the likely sequence of binding, RNA polymerase II entrance upon the scene and subsequent action.
"RNA polymerase II (also called RNAP II and Pol II) ... catalyzes the transcription of DNA to synthesize precursors of mRNA and most snRNA and microRNA.[12] ... In humans RNAP II consists of seventeen protein molecules (gene products encoded by POLR2A-L, where the proteins synthesized from 2C-, E-, and F-form homodimers)."[11]
Preinitiation complexes

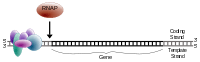
For eukaryotic transcription, the RNA polymerase II holoenzyme de-helicizes the DNA, attaches along the template strand.
Once the preinitiation complex has found its appropriate attachment section along the template strand of DNA, RNA polymerase II is attached and begins transcription.
Start sites
A start site is a biochemically signaled nucleotide or set of nucleotides for attachment either to the Epigenomes or the DNA.
Translation start codon
"Specific sequences of DNA act as a template to synthesize mRNA."[8]
"The start codon is the first codon of an mRNA transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes"[8]
"The most common start codon is AUG [ATG in the DNA]."[8]
"The start codon is almost always preceded by an untranslated region 5' UTR."[8]
For a positive (+) transcription, the start codon on the template strand of DNA is at the end, while a negative (-) transcription has it in the first exon after the 5' UTR.
"Alternate start codons (non ATG) are very rare in eukaryote's nuclear genome. Mitochondrial genomes ... use alternate start codons more significantly (mainly GUG and UUG). For example E. coli uses 83% ATG (AUG), 14% GTG (GUG), 3% TTG (UUG) and one or two others (e.g., ATT and CTG).[13]"[8]
"Note that these alternate start codons are still translated as Met when they are at the start of a protein (even if the codon encodes a different amino acid otherwise). This is because a separate tRNA is used for initiation."[8]
| nonpolar | polar | basic | acidic | (stop codon) |
| 1st base |
2nd base | 3rd base | |||||||
|---|---|---|---|---|---|---|---|---|---|
| T | C | A | G | ||||||
| T | TTT | (Phe/F) Phenylalanine | TCT | (Ser/S) Serine | TAT | (Tyr/Y) Tyrosine | TGT | (Cys/C) Cysteine | T |
| TTC | TCC | TAC | TGC | C | |||||
| TTA | (Leu/L) Leucine | TCA | TAA | Stop (Ochre) | TGA | Stop (Opal) | A | ||
| TTG | TCG | TAG | Stop (Amber) | TGG | (Trp/W) Tryptophan | G | |||
| C | CTT | CCT | (Pro/P) Proline | CAT | (His/H) Histidine | CGT | (Arg/R) Arginine | T | |
| CTC | CCC | CAC | CGC | C | |||||
| CTA | CCA | CAA | (Gln/Q) Glutamine | CGA | A | ||||
| CTG | CCG | CAG | CGG | G | |||||
| A | ATT | (Ile/I) Isoleucine | ACT | (Thr/T) Threonine | AAT | (Asn/N) Asparagine | AGT | (Ser/S) Serine | T |
| ATC | ACC | AAC | AGC | C | |||||
| ATA | ACA | AAA | (Lys/K) Lysine | AGA | (Arg/R) Arginine | A | |||
| ATG[A] | (Met/M) Methionine | ACG | AAG | AGG | G | ||||
| G | GTT | (Val/V) Valine | GCT | (Ala/A) Alanine | GAT | (Asp/D) Aspartic acid | GGT | (Gly/G) Glycine | T |
| GTC | GCC | GAC | GGC | C | |||||
| GTA | GCA | GAA | (Glu/E) Glutamic acid | GGA | A | ||||
| GTG | GCG | GAG | GGG | G | |||||
- The codon ATG both codes for methionine and serves as an initiation site: the first ATG in an mRNA's coding region is where translation into protein begins.[14]
This is the standard or universal genetic code.
This table is found in both DNA Codon Table and Genetic Code. (And probably a few other places.) By default it's the DNA code (using the letter T for Thymine). Use template parameter "T=U" to make it the RNA code (using U for Uracil). See also [Template:Inverse codon table] the inverse codon table.
5'-untranslated region

“The five prime untranslated region (5' UTR), can contain elements for controlling gene expression by way of regulatory elements. It begins at the transcription start site and ends one nucleotide (nt) before the start codon (usually AUG) of the coding region. ... The 5' UTR has a median length of ~150 nt in eukaryotes, but can be as long as several thousand bases. ... Several regulatory sequences may be found in the 5' UTR:
- Binding sites for proteins, that may affect the mRNA's stability or translation, for example iron responsive elements, that regulate gene expression in response to iron.
- Riboswitches.
- Sequences that promote or inhibit translation initiation.
- Introns within 5' UTRs have been linked to regulation of gene expression and mRNA export[15].”[16]
Primer extension
"Primer extension is a technique whereby the 5' ends of RNA or DNA can be mapped."[17]
"Primer extension can be used to determine the start site of RNA transcription for a known gene."[17][18]
"This technique requires a radiolabelled primer (usually 20 - 50 nucleotides in length) which is complementary to a region near the 3' end of the gene. The primer is allowed to anneal to the RNA and reverse transcriptase is used to synthesize cDNA from the RNA until it reaches the 5' end of the RNA. By running the product on a polyacrylamide gel, it is possible to determine the transcriptional start site, as the length of the sequence on the gel represents the distance from the start site to the radiolabelled primer."[17]
A radioisotope used for primer labeling is 32P.[18]
Focused promoters
"In focused transcription, there is either a single major transcription start site or several start sites within a narrow region of several nucleotides. Focused transcription is the predominant mode of transcription in simpler organisms."[7]
"Focused transcription initiation occurs in all organisms, and appears to be the predominant or exclusive mode of transcription in simpler organisms."[7]
"In vertebrates, focused transcription tends to be associated with regulated promoters".[7]
"The analysis of focused core promoters has led to the discovery of sequence motifs such as the TATA box, BREu (upstream TFIIBrecognition element), Inr (initiator), MTE (motif ten element), DPE (downstream promoter element), DCE (downstream core element), and XCPE1 (Xcore promoter element 1) [...]."[7]
Dispersed promoters
"In dispersed transcription, there are several weak transcription start sites over a broad region of about 50 to 100 nucleotides. Dispersed transcription is the most common mode of transcription in vertebrates. For instance, dispersed transcription is observed in about two-thirds of human genes."[7]
In vertebrates, "dispersed transcription is typically observed in constitutive promoters in CpG islands."[7]
Core promoters
"Focused transcription typically initiates within the Inr, and the A nucleotide in the Inr consensus is usually designed as the “+ 1” position, whether or not transcription actually initiates at that particular nucleotide. This convention is useful because other core promoter motifs, such as the MTE and DPE, function with the Inr in a manner that exhibits a strict spacing dependence with the Inr consensus sequence (and hence, the A + 1 nucleotide) rather than the actual transcription start site (Burke and Kadonaga, 1997, Kutach and Kadonaga, 2000 and Lim et al., 2004)."[7]
"With TATA-driven core promoters, transcription can be achieved in vitro with purified RNA polymerase II, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH."[7]
"NC2 (negative cofactor 2; also known as Dr1-Drap1) [...] was identified as repressor of TATA-dependent transcription [...]."[7]
"TBP (TATA box-binding protein) activates TATA transcription [...] The TBP subunit binds to the TATA box [...] TFIIA appears to promote the binding of TBP to the TATA box."[7]
The core promoter is the minimal portion of the promoter required to properly initiate transcription. The core promoter is approximately -34 nt upstream from the TSS.
The TATA box and the initiator element act synergistically when separated by 25-30 nt but act independently when separated by more than 30 nt.[19] When separated by 15 or 20 bp synergy is retained, and the location of the TSS is dictated by the location of the TATA box rather than the location of the Inr.[19]
Research
Hypothesis:
- RNA polymerase II holoenzyme complex looks for or is keyed by DNA nucleotide sequences to the TSS(s).
Control groups

The findings demonstrate a statistically systematic change from the status quo or the control group.
“In the design of experiments, treatments [or special properties or characteristics] are applied to [or observed in] experimental units in the treatment group(s).[20] In comparative experiments, members of the complementary group, the control group, receive either no treatment or a standard treatment.[21]"[22]
Proof of concept
Def. a “short and/or incomplete realization of a certain method or idea to demonstrate its feasibility"[23] is called a proof of concept.
Def. evidence that demonstrates that a concept is possible is called proof of concept.
The proof-of-concept structure consists of
- background,
- procedures,
- findings, and
- interpretation.[24]
See also
References
- ↑ Marketa Zvelebil, Jeremy O. Baum (2008). Understanding bioinformatics. Garland Science. ISBN 978-0815340249.
- ↑ Ino4president (November 4, 2011). "Transcription start site, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. Retrieved 2012-05-16.
- ↑ Nelson, David L. and Michael M. Cox (2008). Principles of Biochemstry, ed. 5, W.H. Freeman and Company, p. 272.
- ↑ "Nitrogenous base, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. March 20, 2014. Retrieved 2014-05-16.
- ↑ "Pyrimidine, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 8, 2014. Retrieved 2014-05-16.
- ↑ "Purine, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. March 30, 2014. Retrieved 2014-05-16.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 Tamar Juven-Gershon and James T. Kadonaga (15 March 2010). "Regulation of gene expression via the core promoter and the basal transcriptional machinery". Developmental Biology 339 (2): 225-9. doi:10.1016/j.ydbio.2009.08.009. http://www.sciencedirect.com/science/article/pii/S0012160609011166. Retrieved 2016-01-16.
- 1 2 3 4 5 6 7 "Start codon, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. October 19, 2012. Retrieved 2012-10-31.
- ↑ Stephen T. Smale and James T. Kadonaga (July 2003). "The RNA Polymerase II Core Promoter". Annual Review of Biochemistry 72 (1): 449-79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. http://www.annualreviews.org/doi/abs/10.1146/annurev.biochem.72.121801.161520. Retrieved 2012-05-07.
- ↑ Myer VE, Young RA (October 1998). "RNA polymerase II holoenzymes and subcomplexes". J. Biol. Chem. 273 (43): 27757–60. doi:10.1074/jbc.273.43.27757. PMID 9774381. http://www.jbc.org/cgi/reprint/273/43/27757.pdf.
- 1 2 "RNA polymerase II holoenzyme, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 1, 2014. Retrieved 2014-05-16.
- ↑ Kornberg R (1999). "Eukaryotic transcriptional control". Trends in Cell Biology 9 (12): M46. doi:10.1016/S0962-8924(99)01679-7. PMID 10611681.
- ↑ Frederick R. Blattner, Guy Plunkett III, Craig A. Bloch, Nicole T. Perna, Valerie Burland, Monica Riley, Julio Collado-Vides, Jeremy D. Glasner, Christopher K. Rode, George F. Mayhew, Jason Gregor, Nelson Wayne Davis, Heather A. Kirkpatrick, Michael A. Goeden, Debra J. Rose, Bob Mau, Ying Shao (September 1997). "The Complete Genome Sequence of Escherichia coli K-12". Science 277 (5331): 1453-62. doi:10.1126/science.277.5331.1453. http://www.sciencemag.org/content/277/5331/1453.
- ↑ Nakamoto T (March 2009). "Evolution and the universality of the mechanism of initiation of protein synthesis". Gene 432 (1–2): 1–6. doi:10.1016/j.gene.2008.11.001. PMID 19056476.
- ↑ Cenik, C, et al. (2011). "Genome analysis reveals interplay between 5' UTR introns and nuclear mRNA export for secretory and mitochondrial genes". PLoS Genetics 7 (4). doi:10.1371/journal.pgen.1001366. http://www.plosgenetics.org/article/info:doi/10.1371/journal.pgen.1001366.
- ↑ "Five prime untranslated region, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 19, 2014. Retrieved 2014-05-16.
- 1 2 3 "Primer extension, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. March 23, 2012. Retrieved 2013-02-14.
- 1 2 John W. Little (August 18, 2010). "Primer extension". Tucson, Arizona USA: University of Arizona. Retrieved 2013-02-14.
- 1 2 A O'Shea-Greenfield, S T Smale (January 15, 1992). "Roles of TATA and initiator elements in determining the start site location and direction of RNA polymerase II transcription". Journal of Biological Chemistry 267 (2): 1391-402. PMID 1730658. http://www.jbc.org/content/267/2/1391.full.pdf+html. Retrieved 2012-05-16.
- ↑ Klaus Hinkelmann, Oscar Kempthorne (2008). Design and Analysis of Experiments, Volume I: Introduction to Experimental Design (2nd ed.). Wiley. ISBN 978-0-471-72756-9. http://books.google.com/?id=T3wWj2kVYZgC&printsec=frontcover.
- ↑ R. A. Bailey (2008). Design of comparative experiments. Cambridge University Press. ISBN 978-0-521-68357-9. http://www.cambridge.org/uk/catalogue/catalogue.asp?isbn=9780521683579.
- ↑ "Treatment and control groups, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 18, 2012. Retrieved 2012-05-31.
- ↑ "proof of concept, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. November 10, 2012. Retrieved 2013-01-13.
- ↑ Ginger Lehrman and Ian B Hogue, Sarah Palmer, Cheryl Jennings, Celsa A Spina, Ann Wiegand, Alan L Landay, Robert W Coombs, Douglas D Richman, John W Mellors, John M Coffin, Ronald J Bosch, David M Margolis (August 13, 2005). "Depletion of latent HIV-1 infection in vivo: a proof-of-concept study". Lancet 366 (9485): 549-55. doi:10.1016/S0140-6736(05)67098-5. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1894952/. Retrieved 2012-05-09.
Further reading
- Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST (January 1994). "DNA Sequence Requirements for Transcriptional Initiator Activity in Mammalian Cells". Mol Cell Biol. 14 (1): 116-27. PMID 8264580. http://mcb.asm.org/cgi/reprint/14/1/116?view=long&pmid=8264580.
- Liston DR, Johnson PJ (March 1999). "Analysis of a Ubiquitous Promoter Element in a Primitive Eukaryote: Early Evolution of the Initiator Element". Mol Cell Biol. 19 (3): 2380-8. http://mcb.asm.org/cgi/content/full/19/3/2380.
- Myer VE, Young RA (October 1998). "RNA polymerase II holoenzymes and subcomplexes". J. Biol. Chem. 273 (43): 27757–60. doi:10.1074/jbc.273.43.27757. PMID 9774381. http://www.jbc.org/cgi/reprint/273/43/27757.pdf.
- Tsvetkov S, Ivanova E, Djondjurov L (December 1989). "The pool of histones in the nucleosol and cytosol of proliferating Friend cells is small, uneven and chasable". Biochem J. 264 (3): 785–91. PMID 2619716. PMC 1133654. //www.ncbi.nlm.nih.gov/pmc/articles/PMC1133654/.
External links
- African Journals Online
- Bing Advanced search
- GenomeNet KEGG database
- Google Books
- Google scholar Advanced Scholar Search
- Home - Gene - NCBI
- JSTOR
- Lycos search
- NCBI All Databases Search
- NCBI Site Search
- PubChem Public Chemical Database
- Questia - The Online Library of Books and Journals
- SAGE journals online
- Scirus for scientific information only advanced search
- SpringerLink
- Taylor & Francis Online
- WikiDoc The Living Textbook of Medicine
- Wiley Online Library Advanced Search
- Yahoo Advanced Web Search
| |||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||
![]() This is a research project at http://en.wikiversity.org
This is a research project at http://en.wikiversity.org
| |
Development status: this resource is experimental in nature. |
| |
Educational level: this is a research resource. |
| |
Resource type: this resource is an article. |
| |
Resource type: this resource contains a lecture or lecture notes. |
| |
Subject classification: this is a biochemistry resource. |
| |
Subject classification: this is a genetics resource. |
| |
Subject classification: this is a medicine resource. |