Gene transcriptions/A1BG
< Gene transcriptions
A1BG is the official symbol for GeneID: 1, "A1BG alpha-1-B glycoprotein [ Homo sapiens ]", in the National Center for Biotechnology Information (NCBI) gene database for gene-specific information. To access information about this gene, use the "Home - Gene - NCBI" external link and enter "1[uid]", without the quotes, or the number one after the "/". The official name of this gene is "alpha-1-B glycoprotein"[1]. "The protein encoded by this gene is a plasma glycoprotein of unknown function. The protein shows sequence similarity to the variable regions of some immunoglobulin supergene family member proteins."[1]
Transcription of eukaryotic genes is believed to be reasonably well understood. Not all the facts are known but phenomenology is probably complete. Each possibility to cause a simple computer program to transcribe the gene for A1BG is to be tested.
Most undergraduate university courses describe transcription, so many of the terms used here to analyze the transcription should be reasonably understood. Some usage of state-of-the-art refereed journal articles is included.
Although there are computer programs being used to predict the effects of gene expression, up-regulation or down-regulation, most of these are quite advanced and are not readily available for students or researchers outside specific universities or companies performing drug research and testing.
Just about any student with access to a computer in various forms is able to write small programs in a variety of computer languages to test for themselves whether or not a specific algorithm works to transcribe this gene.
The transcription start site for A1BG has probably been discovered through the use of reverse transcriptase. The publication demonstrating this has not been found. Further, the product of the gene has been detected, but exactly how the product has its messenger RNA initially transcripted is unspecified and perhaps unknown.
Notations
Notation: let the symbol / replace or.
For example, (A or C or G) becomes (A/C/G).
Notation: let the following table of International Union of Pure and Applied Chemistry (IUPAC) stand for the nucleotides indicated.
| IUPAC nucleotide code | Base |
|---|---|
| A | Adenine |
| B | C/G/T |
| C | Cytosine |
| D | A/G/T |
| G | Guanine |
| H | A/C/T |
| K | G/T |
| M | A/C |
| N | any base (A/C/G/T) |
| R | A/G |
| S | G/C |
| T | Thymine |
| V | A/C/G |
| W | A/T |
| Y | C/T |
Control group
For the transcription of A1BG, a control group would likely have a DNA segment that contains at least one transcription start site, e.g., G/A/T-G/C+1-G-T/C-G-G-G/A-A-G/C. This segment directs the RNA polymerase II holoenzyme to the exact TSS.
In terms of a promoter, this transcription by definition should occur in either the core promoter or the proximal promoter. If not fully understood, the TSS may occur in either a focused promoter or a dispersed promoter.
A transcriptional characteristic of a promoter element may be to be located between -8 and +2 relative to at least one TSS.
Each promoter element may be found in at least one gene or isoform.
Genomes

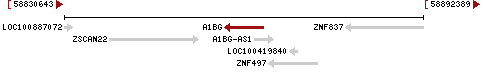
Shown on the top diagram is the genomic context of A1BG as of 2010. The gene is overlapped by a non-coding RNA (NCRNA00181), and has nearby genes ZSCAN22 at the 5' end and ZNF497 (GeneID: 162968) at the 3' end. The nucleotides between genes ZSCAN22 (GeneID: 342945) and the 5'-end of A1BG, should contain the A1BG promoter. NCRNA00181, which overlaps A1BG, has numbered nucleotides 58863336 to 58866548.
In the lower diagram NCRNA00181 has undergone a name change to A1BG antisense RNA 1 (A1BG-AS1), GeneID: 503538.
Genes
Def. a "unit of heredity; a segment of DNA or RNA that is transmitted from one generation to the next, and that carries genetic information such as the sequence of amino acids for a protein"[2] is called a gene.
Gene expressions
Although it is harder to regulate the transcription of genes with multiple transcription start sites, "variations in the expression of a constitutive gene would be minimized by the use of multiple start sites."[3]
Earlier "studies led to the design of a super core promoter (SCP) that contains a TATA, Inr, MTE, and DPE in a single promoter (Juven-Gershon et al., 2006b). The SCP is the strongest core promoter observed in vitro and in cultured cells and yields high levels of transcription in conjunction with transcriptional enhancers. These findings indicate that gene expression levels can be modulated via the core promoter."[3]
Pancreatic juice is "an exceptionally rich source of proteins which are released from pancreatic cells in the physiological state".[4] Matrix metalloproteinase-9 (MMP-9), A1BG, and oncogene DJ-1 occur in the pancreatic cancer juice.[4] "MMP-9, DJ-1 and A1BG [are] positively expressed in 82.4%, 72.5% and 86.3% of pancreatic cancer tissues, significantly higher than that in normal pancreas tissues."[4]
Gene transcriptions
The detection of the gene product presumes that transcription occurs and may suffice as proof of concept.
Def. "the factors, including RNA polymerase II itself, that are minimally essential for transcription in vitro from an isolated core promoter"[5] are called the basal transcription machinery.
RNA polymerase II holoenzyme complex finds and uses the transcription start site. How?
Immunoglobulin domain
"The immunoglobulin domain is a type of protein domain that consists of a 2-layer sandwich of between 7 and 9 antiparallel β-strands arranged in two β-sheets with a Greek key topology.[6][7]"[8]
Angiotensin-converting enzyme
The angiotensin I converting enzyme (ACE) is "involved in catalyzing the conversion of angiotensin I into a physiologically active peptide angiotensin II" and has a "testicular form ... variant (2)".[9]
ACE "plays a key role in the renin-angiotensin system."[9]
"CRISP proteins have been shown to be involved in various functions related to sperm–oocyte fusion, innate host defense function and ion channel blockage."[10]
"Multiple members of the CRISP family have been identified in the mammalian male genital tract (CRISP1, CRISP2 and CRISP3)."[10]
"[T]here is evidence that prostate cancer patients with higher levels of CRISP3 have a smaller probability of recurrence-free outcomes [13]."[11]
A1BG-CRISP3
The human cysteine-rich secretory protein (CRISP3) "is present in exocrine secretions and in secretory granules of neutrophilic granulocytes and is believed to play a role in innate immunity."[12] CRISP3 has a relatively high content in human plasma.[12]
"The A1BG-CRISP-3 complex is noncovalent with a 1:1 stoichiometry and is held together by strong electrostatic forces."[12] "Similar [complex formation] between toxins from snake venom and A1BG-like plasma proteins ... inhibits the toxic effect of snake venom metalloproteinases or myotoxins and protects the animal from envenomation."[12]
"Opossums [such as shown at top of the article] have a remarkably robust immune system, and show partial or total immunity to the venom of rattlesnakes, [Agkistrodon piscivorus] cottonmouths, and other [Crotalinae] pit vipers.[13][14]"[15]
"Crisp3 [is] mainly [expressed] in the salivary glands, pancreas, and prostate."[16] "CRISP3 is highly expressed in the human cauda epididymidis and ampulla of vas deferens (Udby et al. 2005)."[16]
Genomic product

As indicated in the diagram above, there are eight exons (red rectangles) and seven introns (red lines) between the 5' untranslated region (UTR, 5'-UTR) and the 3'-UTR.
When A1BG is transcribed by the RNA polymerase II holoenzyme, the pre-mRNA (messenger RNA, mRNA) consists of eight exons and seven introns which are spliced to yield the final mRNA. A1BG is a minus strand transcription. The included nucleotides as numbered in the human genome go from 3'-58858172 to 58864864-5' inclusive and are so transcribed. When the mRNA template 5'-58858172 to 58864864-3' is used to create the protein, the translation proceeds 3'-58864864 to 58858172-5' rather than 5'-58858172 to 58864864-3'.
Gene transcriptions
Eukaryotes have a double helix of DNA surrounded by an epigenome.
"[A] single strand of DNA [has a positive sense (+)] if an RNA version of the same sequence is translated or translatable into protein. Its complementary strand is called antisense (or negative (-) sense)."[17]
"The two complementary strands of double-stranded DNA (dsDNA) are usually differentiated as the "sense" strand and the "antisense" strand. ... [T]he DNA antisense strand ... serves as the source for the protein code, because, with bases complementary to the DNA sense strand, it is used as a template for the mRNA."[17]
"The only real biological information that is important for labeling strands is the location of the 5' phosphate group and the 3' hydroxyl group because these ends determine the direction of transcription and translation."[17]
From a molecular point of view, a transcription complex may not have an obvious way to choose the "antisense" strand from the "sense" strand. It also may not have an obvious way to chose the direction of transcription once a strand has been chosen as the "antisense" strand.
To choose which strand, then which direction, may require chemical cues. A1BG has two sections of nucleotides between itself and neighboring genes:
- between ZSCAN22 and A1BG and
- between ZNF497 and A1BG.
The NCBI gene database conveniently labels the genomic context with increasing nucleotide numbers in the direction of transcription 3'-5' on the template strand. When viewing the genomic regions, transcripts, and products under tools, click on "Sequence Text View". The database informs you which strand you are looking at (negative strand) or by "Flip Strands" the (positive strand).
Eukaryotic transcription of the A1BG "protein-coding gene is preceded by ...
- decondensation of the locus,
- nucleosome remodeling,
- histone modifications,
- binding of transcriptional activators and coactivators to enhancers and promoters, and
- recruitment of the basal transcription machinery to the core promoter."[5]
Preinitiation complexes
"A stable preinitiation complex can form in vitro on TATA-dependent core promoters by association of the basal factors in the following order: TFIID/TFIIA, TFIIB, RNA polymerase II/TFIIF, TFIIE, and then TFIIH."[5]
Promoters
The A1BG promoter is a region of the DNA adjacent to the gene itself that facilitates gene transcription. Within the overall promoter are response elements which provide a secure initial binding site for the RNA polyermase II holoenzyme.
Positions in the promoter are designated relative to the transcription start site (N+1, TSS), with positions upstream having negative numbers counting back from -1 away from the TSS, for example, -100 is 100 nucleotides (nts) before 3'-58858172 (specifically pre-3' at 58858072).
Focused promoters
"In focused transcription, there is either a single major transcription start site or several start sites within a narrow region of several nucleotides. Focused transcription is the predominant mode of transcription in simpler organisms."[3]
"Focused transcription initiation occurs in all organisms, and appears to be the predominant or exclusive mode of transcription in simpler organisms."[3]
"In vertebrates, focused transcription tends to be associated with regulated promoters".[3]
"The analysis of focused core promoters has led to the discovery of sequence motifs such as the TATA box, BREu (upstream TFIIBrecognition element), Inr (initiator), MTE (motif ten element), DPE (downstream promoter element), DCE (downstream core element), and XCPE1 (Xcore promoter element 1) [...]."[3]
Dispersed promoters
"In dispersed transcription, there are several weak transcription start sites over a broad region of about 50 to 100 nucleotides. Dispersed transcription is the most common mode of transcription in vertebrates. For instance, dispersed transcription is observed in about two-thirds of human genes."[3]
In vertebrates, "dispersed transcription is typically observed in constitutive promoters in CpG islands."[3]
Distal promoters
A distal promoter is a portion of the promoter for a particular gene. This distal sequence upstream of the gene is a region of DNA that may contain additional regulatory elements, often with a weaker influence than the proximal promoter.
"[T]he distal promoter ... can range several thousands of nucleotides upstream of the TSS and contains additional regulatory elements called enhancers and silencers."[18]
AGC boxes
An AGC box has the consensus sequence 3'-AGCCGCC-5' in the direction of transcription. It may also occur as 3'-TCGGCGG-5' in the direction of transcription, or inverted which has been reported: 3'-CCGCCGA-5' and 3'-GGCGGCT-5'. Ideally, each of these four should be tested on each of the four possible transcription directions.
A1BG has four possible transcription directions:
- on the negative strand from ZSCAN22 to A1BG,
- on the positive strand from ZSCAN22 to A1BG,
- on the negative strand from ZNF497 to A1BG, and
- on the positive strand from ZNF497 to A1BG.
For each transcription promoter that interacts directly with RNA polymerase II holoenzyme, the four possible consensus sequences need to be tested on the four possible transcription directions, even though some genes may only be transcribed from the negative strand in the 3'-direction on the transcribed strand.
On both strands from ZSCAN22 to A1BG, the consensus sequence 3'-AGCCGCC-5' in the direction of transcription does not occur, nor does the complementary consensus sequence 3'-TCGGCGG-5' in the same direction.
On both strands from ZNF497 to A1BG, the consensus sequence 3'-AGCCGCC-5' and the complementary consensus sequence 3'-TCGGCGG-5' in the direction of transcription do not occur.
On the negative strand from ZSCAN22 to A1BG, the inverted AGC box 3'-CCGCCGA-5' occurs at 1754 nts (-2706 nts from the TSS). This may be an enhancer of transcription rather than a transcriber.
On the negative strand from ZSCAN22 to A1BG, the complement of the inverted AGC box 3'-GGCGGCT-5' does not occur.
On the positive strand from ZSCAN22 to A1BG, the inverted AGC box 3'-CCGCCGA-5' does not occur.
On the positive strand from ZSCAN22 to A1BG, the complement of the inverted AGC box 3'-GGCGGCT-5' occurs at 1754 nts (-2706 nts from the TSS). This may be an enhancer of transcription rather than a transcriber.
On both strands from ZNF497 to A1BG, the inverted AGC box 3'-CCGCCGA-5' and the complement of the inverted AGC box 3'-GGCGGCT-5' do not occur.
Enhancer boxes
"An E-box (Enhancer Box) is a DNA sequence which usually lies upstream of a gene in a promoter region. It is a transcription factor binding site where the specific sequence of DNA, CANNTG, is recognized by proteins that can bind to it to help initiate its transcription. Once transcription factors bind to promoters, they allow for association of other enzymes which will copy the DNA into mRNA. The consensus sequence for the E-box element is CANNTG, with a palindromic canonical sequence of CACGTG.[19] Transcription factors containing the basic helix-loop-helix protein structural motif typically bind to E-boxes or related variant sequences and enhance transcription of the downstream gene."[20]
Proximal promoters
A 'proximal promoter' is a proximal sequence upstream of the gene, specifically the transcription start site (TSS) of the gene, that tends to contain primary regulatory elements. It is approximately 250 nucleotides (nts) upstream (signified by a negative sign before the number of nucleotides, eg. -250 nts) of the TSS and has specific transcription factor binding sites.
"[T]he proximal promoter [is] a region containing several regulatory elements, which ranges up to a few hundred nucleotides upstream of the TSS"[18].
HY boxes
The hypertrophy region HY box is between -89 and -60 nucleotides (nts) upstream from the transcription start site.[21]
Metal responsive elements
"[T]hree potential metal response elements (MREs) [overlap] the E-boxes in the repeats, (TGCACGT with TGCRCNC being the consensus sequence; 17,18)."[22]
The reproducible consensus sequence seems to be 3'-TGCRCNC-5', specifically 3'-TGC-(A/G)-C-(A/C/G/T)-C-5'.
Core promoters
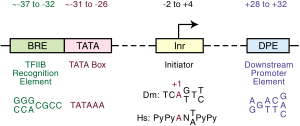
The core promoter is "the minimal portion of the promoter required to properly initiate transcription.[5]"[24] The core promoter is approximately -34 nts upstream from the TSS. "Several factors have been identified that bind to core promoters (reviewed in Smale, 1997)".[25][26]
GC content
Approximately “76% of human core promoters lack TATA-like elements, have a high GC content, and are enriched in Sp1 binding sites.”[27] The core promoter for A1BG does not have a high GC content.
"CpG islands typically occur at or near the transcription start site of genes, particularly housekeeping genes, in vertebrates.[28]"[29]
The number of CG or GC pairs near the TSS for A1BG appears to be low (closer to ~6 % rather than ~60 %).
EIF4E basal elements
The EIF4E basal element, also eIF4E, (4EBE) is a basal promoter element for the eukaryotic translation initiation factor 4E. "Interactions between 4EBE and upstream activator sites are position, distance, and sequence dependent."[30]
The consensus sequence is 3'-TTACCCCCCCTT-5' in the direction of transcription.[30]
CAAT boxes
"[A] CCAAT box (also sometimes abbreviated a CAAT box or CAT box) is a distinct pattern of nucleotides"[31] along the template strand of DNA in eukaryotes.
On the template strand, the CAAT box consensus sequence is 3'-(C/T)(A/G)(A/G)CCAATC(A/G)-5'.
GC boxes
"A GC box sequence, one of the most common regulatory DNA elements of eukaryotic genes, is recognized by the Spl transcription factor; its consensus sequence is represented as 5'-G/T G/A GGCG G/T G/A G/A C/T-3' [or 5′-KRGGCGKRRY-3′] (Briggs et al., 1986)."[32]
B recognition elements
Consensus sequence on the template strand for the B recognition element is 3'-G/C-G/C-G/A-C-G-C-C-5'.
The consensus sequence is 5’-G/C G/C G/A C G C C-3’.[33]
The general consensus sequence using degenerate nucleotides is 5’-SSRCGCC-3’, where S = G or C and R = A or G.[27]
TATA boxes
"The TATA box (also called Goldberg-Hogness box)[34] is a DNA sequence (cis-regulatory element) found in the promoter region of genes in archaea and eukaryotes;[5] approximately 24% of human genes contain a TATA box within the core promoter.[27]".[35]
The TATA box is a "binding site of either general transcription factors or histones"[35].
On the negative strand in the negative transcription direction (from ZSCAN22 to A1BG) there are two TATA boxes: 3'-TATATATA-5' at 1600 (or -2860 nts) and 3'-TATATAAA-5' at 1602 (or -2858 nts). This is way outside the core promoter and may apply to an isoform between these two genes.
On the positive strand in the negative transcription direction there are three TATA boxes: 3'-TATAAAAG-5' at 184 (or -4276 nts), 3'-TATAAAAG-5' at 223 (or -4237 nts), and 3'-TATATAAA-5' at 2874 (or -1586 nts). All of these are way outside the core promoter in the distal promoter at best and most likely relate to one or more isoforms on the positive strand between ZSCAN22 to A1BG.
On both strands in the positive transcription direction from ZNF497 to A1BG there are no TATA boxes.
Complements of and inverted TATA boxes may not have been reported as yet.
Downstream TFIIB recognition
The downstream TFIIB recognition element (dBRE) has a consensus sequence in the transcription direction on the template strand of 3'-RTDKKKK-5', using degenerate nucleotides, or 3'-A/G-T-A/G/T-G/T-G/T-G/T-G/T-5'.[36]
dBRE is cis-TATA box, between the TATA box and the Inr or transcription start site (TSS) and trans-TSS.[36]
X core promoter element 1
The core promoter element X core promoter element 1 (XCPE1) directs activator-, mediator-, protein-dependent but TFIID-independent RNA polymerase II transcription from TATA box-less promoters.[37]
Motif ten elements
The motif ten element (MTE) is a downstream core promoter element that "promotes transcription by RNA polymerase II when it is located precisely at positions +18 to +27 relative to A+1 in the initiator (Inr) element."[38]
GAAC elements
The GAAC element is usually a core promoter element containing guanine (G), adenine (A), and cytosine (C), "able to direct a new transcription start site 2-7 bases downstream of itself, independent of TATA and Inr regions."[39]
Initiator elements
"RNA pol II itself recognizes features of the Inr which might assist the correct positioning of the polymerase on the promoter (Carcamo et al., 1991; Weis and Reinberg, 1997)."[25][40][41]
"[T]he initiator (INR) element located at, or immediately adjacent to, the TSS, ... is recognized by the TBP-associated factors TAF1 and TAF2 of the TFIID complex"[27] "[T]wo [non-Inr] motifs - M3 (SCGGAAGY) and M22 (TGCGCANK) - ... occur preferentially in human TATA-less core promoters."[27]
"[T]ranscription does not need to begin at the +1 nucleotide for the Inr to function. RNA polymerase II has been redirected to alternative start sites by reducing ATP concentrations within a nuclear extract, by altering the spacing between the TATA and Inr in a promoter containing both elements, and by dinucleotide initiation strategies"[5].
The wider consensus sequence of 3'-YYRNWYY-5' allows a G at the TSS but at most only allows two Gs in a row.[42]
The "sharp difference in the specificity of the Inr consensus between Drosophila and mammals suggests that mammalian transcription factors have evolved to function with a broader range of Inr sequences than Drosophila transcription factors. This property may be related to the prevalence of dispersed core promoters in mammals but not in Drosophila."[42]
Angiotensinogen core promoter element
The AGCE1 is supposed to occur before the TSS.[43]
Downstream core elements
The downstream core element (DCE) is a transcription core promoter sequence that is within the transcribed portion of a gene.
The consensus sequence for the DCE is CTTC...CTGT...AGC.[44] These three consensus elements are referred to as subelements: "SI is CTTC, SII is CTGT, and SIII is AGC."[44]
The number of nucleotides between each subelement can apparently vary down to none.
A core promoter that contains all three subelements may be much less common than one containing only one or two.[44] "SI resides approximately from +6 to +11, SII from +16 to +21, and SIII from +30 to +34."[44]
SI as 3'-CTTC-5' can occur as 3 of 4 (CTT, TTC) or 4 of 4 (CTTC). SII as 3'-CTGT-5' can also occur as 3 of 4 (CTG, TGT) or 4 of 4 (CTGT). SIII as AGC is not known to vary.
DCE SIII can function independently of SI and SII.[44]
Downstream promoter elements
"[T]he core sequence of the DPE is located at precisely +28 to +32 relative to the A+1 nucleotide in the Inr"[5]. It is located about 28–33 nucleotides downstream of the transcription start site.[3]
Nucleotides between genes
A1BG is on chromosome 19 between the genes for ZSCAN22 and ZNF497. Before each untranslated region are nucleotides between the genes.
Between ZNF497 and A1BG are 1006 nts before the TSS from this direction. Between ZSCAN22 and A1BG there are 4618 nts.
Depending on which direction the RNA polymerase II holoenzyme transcribes from is the exact nt of the transcription start site (TSS). For transcription from the ZNF497 direction the TSS sits among the local nts as follows: TTGG+1GGC. For transcription from the ZSCAN22 direction, the TSS sits among TGAA+1ACT.
NCBI at the National Institutes of Health includes these in between nucleotides as well as those for each gene. NCBI has predetermined whether the strand is the coding strand (positive strand) or the template strand (negative strand).
The nucleotides between ZNF497 and A1BG as A1BG is approached from ZNF497 on the negative strand are 3'-58865723:
TGAGTGGGGAGGATGCAGTGCAGGGGGCAGATGAGGGCTAGGCGGTTGCCCTGGGCCCTCACACT CGTAGCGGAGCTAGGCTGGGACACCCAGGGTGGGGACCAGACCTCCCCGGGTGGGAATGACAGGA TGTCCATGGAGGCTGAGTGTGAAAGCACCACGGTCTCACCCCTGTCCTGTTCCATCCCAACAGGCT GTGGTGAGGAGAGGGGGAGGCAGGGGAAGCGGGAGGCCTGGCCTCCAGGCAGCAGGCTATAGCCA CATGAGTGACCACCAGCAGCTCAGGTAACTGAGCACATGTCACGGGTAGGCCTGGGAAGCGCAGG TCTCAGCTGAATGACCTGGGTGGAAATCCGACTCCAGAGCCGTGGTGGGTCACACCATGCAGAATG AACCAGTGATGGAGAAGGAACCACAGTCCTCAGGAAGAGTGAGGGTGCACCTCCAGACAGCCCAT GTGAGGGCAACCGCAGAAAGTCTGAAAAGAGGTGAACCCCACCTTTGGTGTCACATGTGCAGTGTG GTGTGACAGGGAGGGGCTCGCTGGGCTTCAGCCCCGGCACTCTCCACTTGACCTCAGCAGCTCCAG GTAGAGTGGGGAGAACTCAGCGTCTCCTTCTAGAACAGGTTCTAGGATCCATCACTGAAATGAGGA TGAGGTGGTTTTAACATCATTTTATCACTCTTGATTTAGTTTATTAATCATACATGATTATTGATTAT AATTGTTGCTGGGCATCCTGAGGCCTCAGAAGTTCACCCTTTGCCCTGACCCCATGGGGGCCCTGC CCCCGCCTTCCGGGAAGGACAAACACGGGAAGAGGTCAGTGCCCGAGCCACCCCACCGCCCTCCC TTGG+1 :58864973-5'.
The nucleotides between ZSCAN22 and A1BG as A1BG is approached from ZSCAN22 on the negative strand are 3'-58853715:
TTTAAGATTGTCTGACTTAAAGAAAAACCTGGTCGGTATACAAGAAATCTTTTCTATGTGGATTTTG TTCCTATACCCTTCAACTCCCGTTTCCCTATCTTTTCCTATATGTTCCATCCGTACTTTGACTTCTTT CGACTACCTCGACGTAATCAGCGCGTTTTGTCTGTAATTCCATATTTTCGTAAAATTCTTAGTACCC CAGTGATACAAAAATATTTTCTTTGTTAGATAATCCTTCTACATTATCAAGTTTTGGTCCGTGTATT ATACTCAGAACATCTTGTCTTAGTGTCACTCTTTGACTGCTTTGGAATTCACATGAACCTTCACGAT TGTGCAAGAAATACTATCTTTTGTAAACTTCTCCGGCCCACGCCACCGAGGGTGGACATTAGGGTC GTGAAACCCTCCGGTTCTGTCCGCCTAGTGCTCCAGTCCTCACGCTCCGTTCGGACCCGTTGTATC ACTTTGACAGATGTTTTTTATGCTTTTAATCGGTCGGACCCGGCCCGTGTCACCTAGTGTGCATTAA GGTCGTGAAACCCTCCGGTTCTGTCCGTCCAGTGCTCCAGTCCTCTAACTCTAGTAGGACCGATTA TACCACACATTGGGGAAGAGATGATTTTTATGTTTTTTAACCGGTCCGTGCCACCGAGTGCGGACA TTAGGGTCGTGAAACCCTCCGGTTCCGTCCGCCTAGTGCTCCAGTCCTCAAGCTCTGGTCGGACTG GTCGCACCACTGTGGGGCAGAGATGATTTTTATGTTTTTTAATCGACCTACACCACCACACATGGAC ATTAGGGTCGATGATTCCTCCGACTCCGTCCTCTTAGCGAACTTGGGTCCTCCGCCTCCAACGCCA CTCGGTTCTAGTGTGGTAACGCGAGGTCGGACCCGTTGTCTCGCTCTGAGACAGAGTTTTTTTTTTT TTTTTTTTTCGGTCCGTACCGCCGTGTGCGGACATCTAGGTCGATTAGTCCTCCGACTCCGTCCTCT TAACGAACTTGGACCCTCCGTCTCCAACGTCACTCGGCTCTAACCCACTGAAGTGAGGTCGGAGCT GTTGTCTCACTCTGAGACAGAGTTTTTTTTTTTTTTTTTCCGACCCGTGTCACCGAGTGTGGACATT AGGGTCGTGAAACCCTGCGGCTCCACCCACCTAGTGGACTCAAGTGCTCAAACGCTGGTCGGACCG GTTGTACCACTTTGGGGCACAGATGTTTTTTAATCGGCCCGCACCACCGCCCACGGACATTAGGGT CGATGAGTCCTCCGACTCCGTCCTCTTAATGAACTTGGATCCTCCGTCTCCAACGTCACTCGGCTC TAACGTGGTAACGTGAGGTCAGACCCGTTATTCTCGTTTTGAGGTAAAGTTTTTGTTTGTTTTTTTT CTGAGTTGGGTCTTAAGAAAAAAAAAAAAAAAAACTCTACCTCAGAGCGAGACAACGGGTCCGACC TCACGTCACCACACTAGAGTCGAGTGACGTTCGAGGCGGAGGGCAGTTGGGTCTTAAGATATAGGT CACGTTTGTGTGAATTTCTTGCTTCCGTTTTATGTCTGTAGGAGTTTACTTTGTTTGGATTCTATATA TAAAGAACGGTCGTCTGAACGGACTTTTCTTTACGGTTTCCTTTGAGAACCATCTTCCCTTTACTAT GGTCTCCCTTTCGCCCTTGAAACCCTTAATTTACTATCATTATCTCTAACGTGTTAATAAAATAGAA ATTTTATACCGACCTCCGCCGACCCGCGCCACCGAGTGCAGACATTAGGGTCGTGAAACCCTCCGA CTCCACCCGCCTAGGGTTCCCGTCCTCTACCTCTGGTAGGACCGATTGTACAACTTTGGGGTAGAG ATGATTTTTATGTTTTTTTAATCGACCCGCACCACCACCCGAGGACATTAGGGTCGATGAACCCTCT GACTCCGTCCTCTTACCGTACTTGGACCCTCCGTCTCGAACGTCACTCGGCTCTAGTGCGGTGACG TGAGGTCGGACCCGCTGTCTCGTTCTGAGACAGAGTTTTTTTTTTTTTTTTTTTCTTTTTTTTACACC GACCTCCGGTCCACGTCACATTAGGATCGTGAAACCCTCCGACTCCACCTGTCTGGTGAACTCGAG TCCTCAAACTCTGGTTGTACCGCTTTATGACAGAGATGATTTCTATGTTTTTTACTGGCCCACGCCA CCGAGTGCGGTCATGAAACCCTCCGACTCCGCCAACCCTAGTGTTCCAGTCCTCAAACTCTGGTCG GACCTGTCGTACCACTTTGGGGTAGAGATGATTTTTATGTTTTTTGATCGGCCCGTACGACCACCCA CGGACATCATGGTCGATGAGGCCTCCGACTCCGTCCTCTTAACGAACTTGGACCGTCCGCCTCCAA CGTCACTCGGCTCTAGTGTGGTGACGTGAGGTCGGACCCGTTGTCTCACTCTAAGCCCTTTTTTTTT TTTTTTCGTTTTCGTTTGTTTGTTTTGAGTTATCATTCTTTTGTTTGTCCGGTCCGTGCCACCGAGTA CGGACATCAGGGTTGTGAAACCTTCCGACTCCGTCCACCTAGTGAACTCCAGTCCTCAACTTCTGG TCACACCGGTTGTACCACTTTGGGGGAGAGGTGATTTATATGTTTTTAGTCGGTCACACCACCGTG TACGGACATTAGGGTCGATGTGTCCTCCGACTCCCTCAACTTAGCGAACTTGGACCCGCCGCCTTC AACGTCACTCGACTTTAGTACGGTGACGTGGGGTCGGACCCGTTGTCTCGTACTGAGAGAGTTTTT CTTTCTTTTCTCTTCTTTTTTCTTTTCTCTTCTTTTTTCTTTTGTTGGGTTATAAAATTTCACACGTTT TATATATTTGTCTGTAAAGTAGTTTCTACGATATATCTACCGTTGATTCGTGAACCCCTTTTTTACGA ATTGTAGTAGTCTGTAATTCCTTTGCGTTCCTTTTGGTGATAATCTATACAGATGTATGGATAATCT TACCGATTTTATTTGAAAAATTTTTTGATTCGACCCCGACCCGCACCACCGAGTGGGGACATTAGGG TCGTGAAACCCTCCGACTCCGCCCACCTAGTGAACTCGAGTCCTCAACCTCTGGTCGGACCGGTGG GTTGTTCCACTTTGGGGTAGAGATGATTTTTGTATTTTTAATCGACCCACACCACCACCCACGGACA GTCGGGTCGATGAGTCCTCCGACTCCGTGCTCTTAGTGAACTTGGGTCCTCCATCTCCAACGACAC TCGGTTCTAGTGCGGTAACGTGAGGTCGGACCCGTTGTTCTCGTTTTAAGACAGAGTTTTTGTTTAT TTGTTCTTTTCCAAAATTTTTAACTATTTGGTGATCGTTCTTTCTCTCTCTCCTGTGTAGTCCGAACT CTCTTCACTGTAGTAGTATCTGACACGTCTGTAATTTTCCGTATCCGTATAATCCTTGATCTATAAT GATTCCGTCATTTGGTCTAGTGATCGTTTAAGCAACTTTTTGTGTTAAGTAGTTGATAACCAACTAT TATGACTATGCGTAGAAGACACACGGTCCCGGAACTCCGGGACGAGGTCCGTCTTTGAGCCGTCAC CAACCCTTCCGTAGTCTACAGTCTACCGTGTTCTCCTGAGTCTCGACTCCCTATTACCCTTTCTTGT GTCTCCTTAGGTCGGTAAAGGTGTCGCAGGTCGAGACGACACCCTCCGACCCTTGTCGGGTCGTGA TGGTGGGACCTGACCCTCCTGTTCTGGTGTTTTACGTCGAAGGGACTTGGAGGAGAACCACTACCC CAACTACACCAGCTCCATCTCCACTCATACAGACCTCGGAGTTCGTTGACGGGTATGGACGACCCT GGTCCGGCTCCACGGGTCCCTCATTCTCCGTCGTAGGACCTTCTCGTGTCTACTTTCTCCGGGACT CTTCACAACCAACCGGTCCACGACACCGAGTGTGGACATTAGGGTTGTGAAACCCTCCGACTCCGC CCTCCTAGTGAACTCGGGTTCTCAAGTTCTGGTCGGACCCGTTGTATCACTCTGAGTAGAGATGTT TTTTATTTATTATCTTTCTTTTTACACTCAACCGGTCCGTACCACCGAGTACGGACATCAGGGTCGA TGAGTCCTCCGACTCCACCCTCCTAGTGAATTCCGGTCCTCAAGTTCTGTTCGAACCCGTTGTGTCA CTCTGGGACAGATGTTTTTTATTATTAATCGGTCTGCACCGCTACGTACGGGTCCGAGGGTCGATG AACCCTCCGACTCCGTCCTCCTAGCGAACTCGGACCCTCCAGTTTTGACGTCACTCGGCCTGACGT TGTGACGTGAGGTCGGACCCACTGTCACACTCTGGGACAGAGTTTTTTCTTTTTTCTTTTCTTTTGA CACGAGAATTCTCGGTCAAGAGGTGAGGAGATGGAGTCCTCGGTGGGGTCTTGGGTAGGTGAA+1 :58858175-5'.
The nucleotides between ZNF497 and A1BG as A1BG is approached from ZNF497 on the positive strand are 3'-58865723:
ACTCACCCCTCCTACGTCACGTCCCCCGTCTACTCCCGATCCGCCAACGGGACCCGGGAGTGTGAG CATCGCCTCGATCCGACCCTGTGGGTCCCACCCCTGGTCTGGAGGGGCCCACCCTTACTGTCCTAC AGGTACCTCCGACTCACACTTTCGTGGTGCCAGAGTGGGGACAGGACAAGGTAGGGTTGTCCGACA CCACTCCTCTCCCCCTCCGTCCCCTTCGCCCTCCGGACCGGAGGTCCGTCGTCCGATATCGGTGTA CTCACTGGTGGTCGTCGAGTCCATTGACTCGTGTACAGTGCCCATCCGGACCCTTCGCGTCCAGAG TCGACTTACTGGACCCACCTTTAGGCTGAGGTCTCGGCACCACCCAGTGTGGTACGTCTTACTTGGT CACTACCTCTTCCTTGGTGTCAGGAGTCCTTCTCACTCCCACGTGGAGGTCTGTCGGGTACACTCCC GTTGGCGTCTTTCAGACTTTTCTCCACTTGGGGTGGAAACCACAGTGTACACGTCACACCACACTGT CCCTCCCCGAGCGACCCGAAGTCGGGGCCGTGAGAGGTGAACTGGAGTCGTCGAGGTCCATCTCAC CCCTCTTGAGTCGCAGAGGAAGATCTTGTCCAAGATCCTAGGTAGTGACTTTACTCCTACTCCACCA AAATTGTAGTAAAATAGTGAGAACTAAATCAAATAATTAGTATGTACTAATAACTAATATTAACAAC GACCCGTAGGACTCCGGAGTCTTCAAGTGGGAAACGGGACTGGGGTACCCCCGGGACGGGGGCGG AAGGCCCTTCCTGTTTGTGCCCTTCTCCAGTCACGGGCTCGGTGGGGTGGCGGGAGGGAACC+1 :58864973-5'.
The nucleotides between ZSCAN22 and A1BG as A1BG is approached from ZSCAN22 on the positive strand are 3'-58853715:
AAATTCTAACAGACTGAATTTCTTTTTGGACCAGCCATATGTTCTTTAGAAAAGATACACCTAAAAC AAGGATATGGGAAGTTGAGGGCAAAGGGATAGAAAAGGATATACAAGGTAGGCATGAAACTGAAGA AAGCTGATGGAGCTGCATTAGTCGCGCAAAACAGACATTAAGGTATAAAAGCATTTTAAGAATCAT GGGGTCACTATGTTTTTATAAAAGAAACAATCTATTAGGAAGATGTAATAGTTCAAAACCAGGCACA TAATATGAGTCTTGTAGAACAGAATCACAGTGAGAAACTGACGAAACCTTAAGTGTACTTGGAAGTG CTAACACGTTCTTTATGATAGAAAACATTTGAAGAGGCCGGGTGCGGTGGCTCCCACCTGTAATCC CAGCACTTTGGGAGGCCAAGACAGGCGGATCACGAGGTCAGGAGTGCGAGGCAAGCCTGGGCAAC ATAGTGAAACTGTCTACAAAAAATACGAAAATTAGCCAGCCTGGGCCGGGCACAGTGGATCACACG TAATTCCAGCACTTTGGGAGGCCAAGACAGGCAGGTCACGAGGTCAGGAGATTGAGATCATCCTGG CTAATATGGTGTGTAACCCCTTCTCTACTAAAAATACAAAAAATTGGCCAGGCACGGTGGCTCACGC CTGTAATCCCAGCACTTTGGGAGGCCAAGGCAGGCGGATCACGAGGTCAGGAGTTCGAGACCAGCC TGACCAGCGTGGTGACACCCCGTCTCTACTAAAAATACAAAAAATTAGCTGGATGTGGTGGTGTGT ACCTGTAATCCCAGCTACTAAGGAGGCTGAGGCAGGAGAATCGCTTGAACCCAGGAGGCGGAGGTT GCGGTGAGCCAAGATCACACCATTGCGCTCCAGCCTGGGCAACAGAGCGAGACTCTGTCTCAAAAA AAAAAAAAAAAAAAAGCCAGGCATGGCGGCACACGCCTGTAGATCCAGCTAATCAGGAGGCTGAGG CAGGAGAATTGCTTGAACCTGGGAGGCAGAGGTTGCAGTGAGCCGAGATTGGGTGACTTCACTCCA GCCTCGACAACAGAGTGAGACTCTGTCTCAAAAAAAAAAAAAAAAAGGCTGGGCACAGTGGCTCAC ACCTGTAATCCCAGCACTTTGGGACGCCGAGGTGGGTGGATCACCTGAGTTCACGAGTTTGCGACC AGCCTGGCCAACATGGTGAAACCCCGTGTCTACAAAAAATTAGCCGGGCGTGGTGGCGGGTGCCTG TAATCCCAGCTACTCAGGAGGCTGAGGCAGGAGAATTACTTGAACCTAGGAGGCAGAGGTTGCAGT GAGCCGAGATTGCACCATTGCACTCCAGTCTGGGCAATAAGAGCAAAACTCCATTTCAAAAACAAA CAAAAAAAAGACTCAACCCAGAATTCTTTTTTTTTTTTTTTTTGAGATGGAGTCTCGCTCTGTTGCCC AGGCTGGAGTGCAGTGGTGTGATCTCAGCTCACTGCAAGCTCCGCCTCCCGTCAACCCAGAATTCT ATATCCAGTGCAAACACACTTAAAGAACGAAGGCAAAATACAGACATCCTCAAATGAAACAAACCT AAGATATATATTTCTTGCCAGCAGACTTGCCTGAAAAGAAATGCCAAAGGAAACTCTTGGTAGAAGG GAAATGATACCAGAGGGAAAGCGGGAACTTTGGGAATTAAATGATAGTAATAGAGATTGCACAATT ATTTTATCTTTAAAATATGGCTGGAGGCGGCTGGGCGCGGTGGCTCACGTCTGTAATCCCAGCACT TTGGGAGGCTGAGGTGGGCGGATCCCAAGGGCAGGAGATGGAGACCATCCTGGCTAACATGTTGAA ACCCCATCTCTACTAAAAATACAAAAAAATTAGCTGGGCGTGGTGGTGGGCTCCTGTAATCCCAGC TACTTGGGAGACTGAGGCAGGAGAATGGCATGAACCTGGGAGGCAGAGCTTGCAGTGAGCCGAGA TCACGCCACTGCACTCCAGCCTGGGCGACAGAGCAAGACTCTGTCTCAAAAAAAAAAAAAAAAAAA GAAAAAAAATGTGGCTGGAGGCCAGGTGCAGTGTAATCCTAGCACTTTGGGAGGCTGAGGTGGACA GACCACTTGAGCTCAGGAGTTTGAGACCAACATGGCGAAATACTGTCTCTACTAAAGATACAAAAA ATGACCGGGTGCGGTGGCTCACGCCAGTACTTTGGGAGGCTGAGGCGGTTGGGATCACAAGGTCAG GAGTTTGAGACCAGCCTGGACAGCATGGTGAAACCCCATCTCTACTAAAAATACAAAAAACTAGCC GGGCATGCTGGTGGGTGCCTGTAGTACCAGCTACTCCGGAGGCTGAGGCAGGAGAATTGCTTGAAC CTGGCAGGCGGAGGTTGCAGTGAGCCGAGATCACACCACTGCACTCCAGCCTGGGCAACAGAGTG AGATTCGGGAAAAAAAAAAAAAAAGCAAAAGCAAACAAACAAAACTCAATAGTAAGAAAACAAACA GGCCAGGCACGGTGGCTCATGCCTGTAGTCCCAACACTTTGGAAGGCTGAGGCAGGTGGATCACTT GAGGTCAGGAGTTGAAGACCAGTGTGGCCAACATGGTGAAACCCCCTCTCCACTAAATATACAAAA ATCAGCCAGTGTGGTGGCACATGCCTGTAATCCCAGCTACACAGGAGGCTGAGGGAGTTGAATCGC TTGAACCTGGGCGGCGGAAGTTGCAGTGAGCTGAAATCATGCCACTGCACCCCAGCCTGGGCAACA GAGCATGACTCTCTCAAAAAGAAAGAAAAGAGAAGAAAAAAGAAAAGAGAAGAAAAAAGAAAACAA CCCAATATTTTAAAGTGTGCAAAATATATAAACAGACATTTCATCAAAGATGCTATATAGATGGCAA CTAAGCACTTGGGGAAAAAATGCTTAACATCATCAGACATTAAGGAAACGCAAGGAAAACCACTAT TAGATATGTCTACATACCTATTAGAATGGCTAAAATAAACTTTTTAAAAAACTAAGCTGGGGCTGGG CGTGGTGGCTCACCCCTGTAATCCCAGCACTTTGGGAGGCTGAGGCGGGTGGATCACTTGAGCTCA GGAGTTGGAGACCAGCCTGGCCACCCAACAAGGTGAAACCCCATCTCTACTAAAAACATAAAAATT AGCTGGGTGTGGTGGTGGGTGCCTGTCAGCCCAGCTACTCAGGAGGCTGAGGCACGAGAATCACTT GAACCCAGGAGGTAGAGGTTGCTGTGAGCCAAGATCACGCCATTGCACTCCAGCCTGGGCAACAAG AGCAAAATTCTGTCTCAAAAACAAATAAACAAGAAAAGGTTTTAAAAATTGATAAACCACTAGCAAG AAAGAGAGAGAGGACACATCAGGCTTGAGAGAAGTGACATCATCATAGACTGTGCAGACATTAAAA GGCATAGGCATATTAGGAACTAGATATTACTAAGGCAGTAAACCAGATCACTAGCAAATTCGTTGAA AAACACAATTCATCAACTATTGGTTGATAATACTGATACGCATCTTCTGTGTGCCAGGGCCTTGAGG CCCTGCTCCAGGCAGAAACTCGGCAGTGGTTGGGAAGGCATCAGATGTCAGATGGCACAAGAGGAC TCAGAGCTGAGGGATAATGGGAAAGAACACAGAGGAATCCAGCCATTTCCACAGCGTCCAGCTCTG CTGTGGGAGGCTGGGAACAGCCCAGCACTACCACCCTGGACTGGGAGGACAAGACCACAAAATGCA GCTTCCCTGAACCTCCTCTTGGTGATGGGGTTGATGTGGTCGAGGTAGAGGTGAGTATGTCTGGAG CCTCAAGCAACTGCCCATACCTGCTGGGACCAGGCCGAGGTGCCCAGGGAGTAAGAGGCAGCATC CTGGAAGAGCACAGATGAAAGAGGCCCTGAGAAGTGTTGGTTGGCCAGGTGCTGTGGCTCACACCT GTAATCCCAACACTTTGGGAGGCTGAGGCGGGAGGATCACTTGAGCCCAAGAGTTCAAGACCAGCC TGGGCAACATAGTGAGACTCATCTCTACAAAAAATAAATAATAGAAAGAAAAATGTGAGTTGGCCA GGCATGGTGGCTCATGCCTGTAGTCCCAGCTACTCAGGAGGCTGAGGTGGGAGGATCACTTAAGGC CAGGAGTTCAAGACAAGCTTGGGCAACACAGTGAGACCCTGTCTACAAAAAATAATAATTAGCCAG ACGTGGCGATGCATGCCCAGGCTCCCAGCTACTTGGGAGGCTGAGGCAGGAGGATCGCTTGAGCCT GGGAGGTCAAAACTGCAGTGAGCCGGACTGCAACACTGCACTCCAGCCTGGGTGACAGTGTGAGAC CCTGTCTCAAAAAAGAAAAAAGAAAAGAAAACTGTGCTCTTAAGAGCCAGTTCTCCACTCCTCTACC TCAGGAGCCACCCCAGAACCCATCCACTT+1 :58858175-5'.
According to one source, A1BG is transcribed from the direction of ZNF497: 3' - 58864890: CGAGCCACCCCACCGCCCTCCCTTGG+1GGCCTCATTGCTGCAGACGCTCACCCCAG ACACTCACTGCACCGGAGTGAGCGCGACCATCATG : 58866601-5',[45] where the second 'G' at left of four Gs in a row is the TSS.[46]
To obtain the nts between genes as well as the first several nucleotides within the first UTR, input the GeneID followed by [uid]. Under "Genomic context" in parentheses is (58858172..58864865, complement). These are the nucleotide numbers on the chromosome. The nearest neighboring gene ZNF497 has (58865723..58874214), where 58864801 to 58866601 (see above) are the nucleotides between the genes that contain the promoter for A1BG.
Transcription start sites
Notation: let the symbol PPP denote a promoter prediction program.
Notation: let the symbol CpG stand for cytosine - phosphodiester bond - guanine, which indicates that C and G are next to each other on the same DNA strand connected by phosphate.
"One important question is what the different PPPs are actually trying to predict. Some programs aim to predict the exact location of the promoter region of known protein-coding genes, while others focus on finding the transcription start site (TSS)."[47] "Recent research has shown that there is often no single TSS, but rather a whole transcription start region (TSR) containing multiple TSSs that are used at different frequencies (Frith et al., 2008)."[47] "The most recent large-scale validation of PPPs included more programs than any of the earlier studies and introduced for the first time an evaluation based on all experimentally determined TSSs in the human genome (Abeel et al., 2008a, 2008b)."[47] "[T]he current state-of-the-art in promoter prediction is biased toward housekeeping genes that contain CpG islands."[47]
Each of the currently described promoter elements is tested for possible occurrences between ZSCAN22 and A1BG on both the negative and positive strands, and between ZNF497 and A1BG on both strands, going from the neighboring gene toward A1BG.
E boxes
A1BG has an E-box: 3'-CACATG-5' ending at -2118 nt (G) in the distal promoter region.
MREs
On the negative strand going from ZSCAN22 to A1BG, there are no MREs.
eIF4Es
A1BG does not have an eIF4E basal element on the template strand between ZSCAN22 and A1BG TSS.
HY boxes
The only HY box upstream or downstream from A1BG ends (G) at -3711 nts (3'-TGTGGG-5') upstream from the TSS.
CAAT boxes
A1BG does not have a CAAT box in the transcription direction along the negative strand from ZSCAN22 to A1BG.
GC boxes
A1BG does not have a GC box in the transcription direction on either the positive or negative strand (ZSCAN22 to A1BG).
BREs
There are three B recognition elements (BREs) going along the negative strand from ZSCAN22 to A1BG: 3'-CCACGCC-5' out 380 nts from the last nt of the ending untranslated region for ZSCAN22, 3'-CCGCGCC-5' out 1762, and 3'-CCACGCC-5' out 2197 nts.
"The position in nucleotides (nts) relative to the transcription start site (TSS, +1)" is -35 for the BRE."[27] None of the three BREs located are anywhere near the TSS at some 4600 nts out from ZSCAN22.
TATA boxes
On the positive strand, in the nucleotide region between gene ZSCAN22 (NCBI GeneID: 342945) and A1BG (NCBI GeneID: 1) are 211 TATA box-like 8 nt long sequences. Of these,
- TATAAAAG occurs at 58853713 + 183 nts and
- TATAAAAG at 58853713 + 222. This is a TATA box found with some genes.[48] But, the optimal TBP recognition sequence 3'-TATATAAG-5',[49] does not occur.
- TATATAAA occurs only once at 2874 nts from the end of ZSCAN22. TBP is bound to this sequence and TATAAAAG above.[50][51]
- TATAAA occurs seven times, with the closest one at 2874 nts from the end of ZSCAN22. "In virtually every RNA polymerase II-transcribed gene examined, the sequence TATAAA was present 25 to 30 nts upstream of the transcription start site."[5]
A1BG does not have a TATA box in the core promoter region. There is the sequence 3'-TGCTATATAGATGGCAACTAAGCACTTGGGGAAAAAA-5' for which the first nt (T) is number 58856598 or 1574 nt upstream from the beginning of the 3'-UTR at 58858172. Unless another variant exists, -1574 nt from the beginning of the 3'-UTR is a large number of nts away from the TSS.
The closest TATA box-like sequence is 3'-CTCTTAAG-5' on the template strand at 4408 nts from the end of ZSCAN22, which is upstream from the core promoter.
The extra TATA boxes between ZSCAN22 and A1BG strongly suggest that there is at least one gene (or pseudogene) between ZSCAN22 and A1BG not currently in the NCBI database.
On the negative strand between ZNF497 and A1BG, there are no TATA boxes of the form 3’-TATA-A/T-A-A/T-A/G-5’.
For the negative strand going from ZSCAN22 to A1BG there are two TATA boxes: 3'-TATATATA-5' at 1600 nts and 3'-TATATAAA-5' at 1602 nts. These are way too far from the possible TSS in this direction.
These two TATA boxs in the distal promoter at approximately -2860 nts from the TSS, which suggests that there may be a short gene between ZSCAN22 and A1BG.
dBREs
A1BG does not have a TATA box. The closest 7 nucleotide dBRE is at -450 nts which is outside the core promoter in the distal promoter. The closest 6 nt dBRE of consensus sequence 3'-A/G-T-A/G/T-G/T-G/T-G/T-5' is also at -451 nts.
Consensus sequence 3'-T-A/G/T-G/T-G/T-G/T-G/T-5' has an expression at -108 nts with 3'-TGGGTG-5' in the proximal promoter.
Consensus sequence 3'-A/G-T-A/G/T-G/T-G/T-5' has one dBRE at -99 nts with 3'-GTGTG-5' in the proximal promoter.
3'-T-A/G/T-G/T-G/T-G/T-5' is another 5 nts consensus sequence that has a dBRE at -109 nts with 3'-TGGGT-5'.
Another 5 nts consensus sequence that has the same dBRE at -99 nts is 3'-GTGTG-5' in the proximal promoter.
A 4 nts consensus sequence has a dBRE at -100 nts is 3'-GTGT-5' also in the proximal promoter.
A second set of 4 nts consensus sequence dBREs are at -59 nts and +2 nts. The dBRE has not been reported to include the TSS.
A third 4 nts consensus sequence dBRE occurs at -43 nts, roughly just outside the core promoter, with another including the TSS and none in between.
XCPE 1s
There is no X core promoter element 1 between ZSCAN22 and A1BG on the template strand.
MTEs
There is no motif ten element between ZSCAN22 and A1BG on the template strand.
GAACs
There are two GAAC elements in the distal promoter between ZSCAN22 and A1BG.
A1BG does not have a GAAC element within 2-7 nucleotides of the TSS. The closest GAAC element, 3'-GAACT-5', is -999 nts from the TSS.
Inrs
Along the negative strand from ZSCAN22 to A1BG there are at least 43 initiator elements, with the closest one 3'-TCACACT-5' ending at 4361 nts rather than including the TSS of A+1 at 4460 nts.
In the sequence along the positive strand of nucleotides from genes ZSCAN22 and A1BG, the sequence 3'-CCATCCACT-5' occurs only once, just before the transcription start site (TSS) of the A1BG gene. The DCE 3'-CTT-5' contains the TSS at its 5' end.
The TSS for A1BG has the following nts around it: 3'-CCATCCACTT+1TGAGGACAC-5'. Most genes studied early on contained an adenosine (A+1) at the TSS, a cytosine (C-1), and a few pyrimidines (Pys) surrounding these nts.[52]
Usually the Inr contains the TSS. The sequence 3'-CCACTT+1T-5' does contain the TSS but not at A+1. The nearest other Inr ends -24 nt upstream from the TSS.
There are at least 15 Inrs between ZSCAN22 and A1BG.
The sequence 3'-CCACTT+1T-5' is also an Inr, where the TSS is indicated. But, the only DPE nearby 3'-GGACA-5' begins on the fourth nt (+5) after the TSS (+1), not precisely +28 to +32 relative to the TSS nucleotide. No other DPE is even close to this +28 to +32 window.
AGCE1s
A1BG contains 3'-CTT+1-5'. This is an angiotensinogen core promoter element 1 (AGCE1).
The AGCE1 occurs at 3'-CTT+1-5' and 3'-ATC-5' which ends 5 nts upstream from the first and is the only AGCE1 within -25 and -1 nts of the TSS.
DCEs
The downstream core element (DCE) SI 3'-CTT+1-5' contains the TSS but is not downstream of the TSS. Of the DCE SI elements, the closest is some 60 nts downstream from the TSS (3'-CTTC-5' ending at +69).
Of the DCE SII elements such as 3'-CTGT-5', there are none within +1 to +100 nts downstream. Element 3'-CTG-5' occurs starting at +32 nts, which is some +11 nts further downstream than expected and falls within the range for SIII. The element 3’-TGT-5’ has no occurrence within the TSS and +100 nts downstream.
DCE SIII elements (3’-AGC-5’) occur at +19 and +28 nts. The second DCE SIII is close but overlaps any likely nts for DCE SII and is supposed to be after any DCE SII. The DCE SII is unlikely to be used but the second DCE SIII may be close enough to act alone.
Along the negative strand from ZSCAN22 to A1BG and past the TSS, there are no DCE SIs of the type 3'-CTTC-5' past the TSS. Of the type 3'-CTT-5', only one occurs at 4552 nts or 92 nts past the TSS. For type 3'-TTC-5' of DCE SI the closest past is 3'-TTC-5' ending at 4504, or 44 nts past the TSS.
Type SII 3'-CTGT-5' has the closest one ending at 4468 nts and the next ending at 4507 nts.
DPEs
Within the nucleotides of the negative strand going from gene ZSCAN22 to A1BG are at least 163 downstream promoter elements (DPEs), when using the minimal five-nucleotide consensus sequence. There are three DPEs near the required +28 (4487) to +32 (4491) nts from the TSS at 4460 nts from the end of ZSCAN22: 3'-GGTCG-5' at 4480, 3'-AGTCG-5' at 4489, and 3'-GGACC-5' at 4494 nts.
TAFs
Each of the foregoing core promoter elements does not appear to be involved in the transcription of A1BG. The Inr does not contain the known TSS. The DCE is not supposed to contain the TSS, nor is AGCE1 although it is one nt off by containing it. Currently, the only way to default transcribe A1BG is by directing the transcription program directly to the known TSS.
When there is no TATA box in the promoter, a TAF binds sequence specifically, and forces the TBP to bind non-sequence specifically.
5'-untranslated region

The 5' UTR for A1BG contains some 216 nts, depending upon the location of the TSS.
3’-T+1TGAGGACACGAGATCCCAGCCCACTCAGCCCTGGGAGTCCAAAGACATTTTAAACAGAGCCTCTCTTCACATTTA TTAATTCCTGGGAGGAATGAGGGAGGCTTCTCCAGCCCCCCAGAGACCCCGGCCTTGTGCTGCAACAGGAGGGGA GGGAGCCAGTCCAGAATCCCCGGCACTTCTGAGGACACCAACAGCACCCTGGGCCCGCGGCTGCA-5’
“The five prime untranslated region (5' UTR), can contain elements for controlling gene expression by way of regulatory elements. It begins at the transcription start site and ends one nucleotide (nt) before the start codon (usually AUG) of the coding region. ... The 5' UTR has a median length of ~150 nt in eukaryotes, but can be as long as several thousand bases. ... Several regulatory sequences may be found in the 5' UTR:
- Binding sites for proteins, that may affect the mRNA's stability or translation, for example iron responsive elements, that regulate gene expression in response to iron.
- Riboswitches.
- Sequences that promote or inhibit translation initiation.
- Introns within 5' UTRs have been linked to regulation of gene expression and mRNA export[53].”[54]
TBP binding
If TBP can bind to any variety of seven A/Ts before the TSS, then the sequence 3'-AAAAAATAATAATTA-5' is likely to be the 3'-"xod-ATAT"-5', rather than the traditional 3'-"TATA-box"-5'. The first (T)-5' is at 58858405, only 233 nts from TSS, but way outside the core promoter.
RNA polymerase II holoenzymes
"RNA polymerase II ... is recruited to the promoters of protein-coding genes in living cells.[55]"[56] Or, transcription factories are present and the euchromatin is brought within the nearest transcription factory and A1BG messenger RNA (mRNA) is transcribed.
For those circumstances in which the holoenzyme is built onto the euchromatin, it is necessary to consider the holoenzyme components and the likely sequence of binding, RNA polymerase II entrance upon the scene and subsequent action.
"RNA polymerase II (also called RNAP II and Pol II) ... catalyzes the transcription of DNA to synthesize precursors of mRNA and most snRNA and microRNA.[57] ... In humans RNAP II consists of seventeen protein molecules (gene products encoded by POLR2A-L, where the proteins synthesized from 2C-, E-, and F-form homodimers)."[56]
Research
Hypotheses:
- A1BG may be transcribed from either direction.
- A1BG may be transcribed from either strand.
Control groups

The findings demonstrate a statistically systematic change from the status quo or the control group.
“In the design of experiments, treatments [or special properties or characteristics] are applied to [or observed in] experimental units in the treatment group(s).[58] In comparative experiments, members of the complementary group, the control group, receive either no treatment or a standard treatment.[59]"[60]
Proof of concept
Def. a “short and/or incomplete realization of a certain method or idea to demonstrate its feasibility"[61] is called a proof of concept.
Def. evidence that demonstrates that a concept is possible is called proof of concept.
The proof-of-concept structure consists of
- background,
- procedures,
- findings, and
- interpretation.[62]
See also
- 5' cap
- Chromosome 19
- Enhancer
- Eukaryotic transcription
- General transcription factor II D
- Initiator motif
- Preinitiation complex
- RNA polymerase II holoenzyme complex
- Transcription factor
References
- 1 2 NCBI (November 16 2011). "A1BG alpha-1-B glycoprotein [ Homo sapiens ] - Gene -NCBI". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2011-11-28.
- ↑ "gene, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. 31 January 2015. Retrieved 2015-02-21.
- 1 2 3 4 5 6 7 8 9 Tamar Juven-Gershon and James T. Kadonaga (15 March 2010). "Regulation of gene expression via the core promoter and the basal transcriptional machinery". Developmental Biology 339 (2): 225-9. doi:10.1016/j.ydbio.2009.08.009. http://www.sciencedirect.com/science/article/pii/S0012160609011166. Retrieved 2016-01-16.
- 1 2 3 Mei Tian, Ya-Zhou Cui, Guan-Hua Song, Mei-Juan Zong, Xiao-Yan Zhou, Yu Chen and Jin-Xiang Han (August 2008). "Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients". BMC Cancer 16 (8): 241. PMID 18706098. http://www.biomedcentral.com/1471-2407/8/241. Retrieved 2011-11-28.
- 1 2 3 4 5 6 7 8 Stephen T. Smale and James T. Kadonaga (July 2003). "The RNA Polymerase II Core Promoter". Annual Review of Biochemistry 72 (1): 449-79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. http://www.lps.ens.fr/~monasson/Houches/Kadonaga/CorePromoterAnnuRev2003.pdf. Retrieved 2012-05-07.
- ↑ Bork P, Holm L, Sander C (September 1994). "The immunoglobulin fold. Structural classification, sequence patterns and common core". Journal of Molecular Biology 242 (4): 309–20. doi:10.1006/jmbi.1994.1582. PMID 7932691.
- ↑ Brümmendorf T, Rathjen FG (1995). "Cell adhesion molecules 1: immunoglobulin superfamily". Protein Profile 2 (9): 963–1108. PMID 8574878.
- ↑ "Immunoglobulin domain, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. June 29, 2012. Retrieved 2012-07-15.
- 1 2 ncbi (February 19, 2012). "ACE angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 [ Homo sapiens ]". Bethesda, Maryland, USA: National Institutes of Health. Retrieved 2012-02-26.
- 1 2 Edda Topfer-Petersen, Mahnaz Ekhlasi-Hundrieser, Christiane Kirchhoff, Tosso Leeb, Harald Sieme (October 2005). "The role of stallion seminal proteins in fertilisation". Animal Reproduction Science 89 (1-4): 159-70. doi:10.1016/j.anireprosci.2005.06.018. http://www.sciencedirect.com/science/article/pii/S0378432005001958. Retrieved 2012-02-26.
- ↑ Lucy J. Schmidt, Kevin M. Regan, S. Keith Anderson, Zhifu Sun, Karla V. Ballman, Donald J. Tindall (December 2009). "Effects of the 5 alpha‐reductase inhibitor dutasteride on gene expression in prostate cancer xenografts". The Prostate 69 (16): 1730-43. doi:10.1002/pros.21022. PMID 19676081. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2783419/pdf/nihms138639.pdf. Retrieved 2012-02-26.
- 1 2 3 4 Udby L, Sørensen OE, Pass J, Johnsen AH, Behrendt N, Borregaard N, Kjeldsen L. (October 2004). "Cysteine-rich secretory protein 3 is a ligand of alpha1B-glycoprotein in human plasma". Biochemistry 43 (40): 12877-86. doi:10.1021/bi048823e. PMID 15461460.
- ↑ "The Opossum: Our Marvelous Marsupial, The Social Loner". Wildlife Rescue League.
- ↑ Journal Of Venomous Animals And Toxins – Anti-Lethal Factor From Opossum Serum Is A Potent Antidote For Animal, Plant And Bacterial Toxins. Retrieved 2009-12-29.
- ↑ "Opossum, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. February 3, 2013. Retrieved 2013-02-06.
- 1 2 B Haendler, J Krätzschmar, F Theuring and W D Schleuning (July 1993). "Transcripts for cysteine-rich secretory protein-1 (CRISP-1; DE/AEG) and the novel related CRISP-3 are expressed under androgen control in the mouse salivary gland.". Endocrinology 133 (1): 192-8. doi:10.1210/en.133.1.192. PMID 8319566. http://endo.endojournals.org/content/133/1/192.full.pdf+html. Retrieved 2012-02-20.
- 1 2 3 "Sense (molecular biology), In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. February 12, 2013. Retrieved 2013-02-14.
- 1 2 Thomas Abeel, Yvan Saeys, Eric Bonnet, Pierre Rouzé, and Yves Van de Peer (February 2008). "Generic eukaryotic core promoter prediction using structural features of DNA". Genome Research 18 (2): 310-23. doi:10.1101/gr.6991408. http://genome.cshlp.org/content/18/2/310.full.pdf+html. Retrieved 2012-04-04.
- ↑ Jaideep Chaudhary, Michael K. Skinner. "Basic Helix-Loop-Helix Proteins Can Act at the E-Box within the Serum Response Element of the c-fos Promoter to Influence Hormone-Induced Promoter Activation in Sertoli Cells". Molecular Endocrinology 12 (5): 774–786. http://mend.endojournals.org/cgi/content/full/13/5/774.
- ↑ "E-box, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 13, 2013. Retrieved 2013-04-17.
- ↑ Akiro Higashikawa, Taku Saito, Toshiyuki Ikeda, Satoru Kamekura, Naohiro Kawamura, Akinori Kan, Yasushi Oshima, Shinsuke Ohba, Naoshi Ogata, Katsushi Takeshita, Kozo Nakamura, Ung-Il Chung, Hiroshi Kawaguchi (January 2009). "Identification of the core element responsive to runt-related transcription factor 2 in the promoter of human type x collagen gene". Arthritis & Rheumatism 60 (1): 166-78. doi:10.1002/art.24243. PMID 19116917. http://onlinelibrary.wiley.com/doi/10.1002/art.24243/full. Retrieved 2013-06-18.
- ↑ Barbara Levinson, Rebecca Conant, Rhonda Schnur, Soma Das, Seymour Packman and Jane Gitschier (1996). "A Repeated Element in the Regulatory Region of the MNK Gene and Its Deletion in A Patient With Occipital Horn Syndrome". Human Molecular Genetics 5 (11): 1737-42. doi:10.1093/hmg/5.11.1737. http://hmg.oxfordjournals.org/content/5/11/1737.full. Retrieved 2013-04-15.
- 1 2 Jennifer E.F. Butler, James T. Kadonaga (October 15, 2002). "The RNA polymerase II core promoter: a key component in the regulation of gene expression". Genes & Development 16 (20): 2583–292. doi:10.1101/gad.1026202. PMID 12381658. http://genesdev.cshlp.org/content/16/20/2583.full.
- ↑ "Promoter (genetics), In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. January 29, 2013. Retrieved 2013-02-12.
- 1 2 Gillian E. Chalkley and C. Peter Verrijzer (September 1, 1999). "DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the Initiator". The EMBO Journal 18 (17): 4835-45. PMID 10469661. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1171555/pdf/004835.pdf. Retrieved 2012-04-26.
- ↑ S. T. Smale (1997). "Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes". Biochim. Biophys. Acta. 1351: 73-88.
- 1 2 3 4 5 6 Chuhu Yang, Eugene Bolotin, Tao Jiang, Frances M. Sladek, Ernest Martinez. (March 7, 2007). "Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters". Gene 389 (1): 52-65. doi:10.1016/j.gene.2006.09.029. PMID 17123746. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1955227/?tool=pubmed.
- ↑ Saxonov S, Berg P, Brutlag DL (2006). "A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters". Proc Natl Acad Sci USA 103 (5): 1412–1417. doi:10.1073/pnas.0510310103. PMID 16432200. PMC 1345710. //www.ncbi.nlm.nih.gov/pmc/articles/PMC1345710/.
- ↑ "CpG site, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 15, 2014. Retrieved 2014-05-16.
- 1 2 Mary Lynch, Li Chen, Michael J. Ravitz, Sapna Mehtani, Kevin Korenblat, Michael J. Pazin and Emmett V. Schmidt (August 2005). "hnRNP K Binds a Core Polypyrimidine Element in the Eukaryotic Translation Initiation Factor 4E (eIF4E) Promoter, and Its Regulation of eIF4E Contributes to Neoplastic Transformation". Molecular and Cellular Biology 25 (15): 6436-53. doi:10.1128/MCB.25.15.6436-6453.2005. http://mcb.asm.org/content/25/15/6436.full. Retrieved 2013-03-17.
- ↑ "CAAT box, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 8, 2013. Retrieved 2013-04-14.
- ↑ H Imataka, K Sogawa, KI Yasumoto, Y Kikuchi, K Sasano, A Kobayashi, M Hayami, and Y Fujii-Kuriyama (October 1992). "Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene". The EMBO Journal 11 (10): 3663-71. PMID 1356762. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC556826/pdf/emboj00095-0180.pdf. Retrieved 2013-01-27.
- ↑ Alan K. Kutach, James T. Kadonaga (July 2000). "The Downstream Promoter Element DPE Appears To Be as Widely Used as the TATA Box in Drosophila Core Promoters". Molecular and Cellular Biology 20 (13): 4754-64. PMID 10848601. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC85905/pdf/mb004754.pdf. Retrieved 2012-07-15.
- ↑ Lifton RP, Goldberg ML, Karp RW, Hogness DS (1978). "The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications". Cold Spring Harb Symp Quant Biol 42: 1047–51. PMID 98262.
- 1 2 "TATA box, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 30, 2012. Retrieved 2012-05-10.
- 1 2 Wensheng Deng, Stefan G.E. Roberts (October 15, 2005). "A core promoter element downstream of the TATA box that is recognized by TFIIB". Genes & Development 19 (20): 2418–23. doi:10.1101/gad.342405. PMID 16230532. http://genesdev.cshlp.org/content/19/20/2418.full.
- ↑ Yumiko Tokusumi, Ying Ma, Xianzhou Song, Raymond H. Jacobson, and Shinako Takada (March 2007). "The New Core Promoter Element XCPE1 (X Core Promoter Element 1) Directs Activator-, Mediator-, and TATA-Binding Protein-Dependent but TFIID-Independent RNA Polymerase II Transcription from TATA-Less Promoters". Molecular and Cellular Biology 27 (5): 1844-58. doi:10.1128/MCB.01363-06. PMID 17210644. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1820453/. Retrieved 2013-02-09.
- ↑ Chin Yan Lim, Buyung Santoso, Thomas Boulay, Emily Dong, Uwe Ohler, and James T. Kadonaga (July 1, 2004). "The MTE, a new core promoter element for transcription by RNA polymerase II". Genes & Development 18 (13): 1606-17. doi:10.1101/gad.1193404. PMID 15231738. http://genesdev.cshlp.org/content/18/13/1606.full. Retrieved 2013-02-10.
- ↑ Upinder Singh, Joshua B. Rogers (August 21, 1998). "The Novel Core Promoter Element GAAC in the hgl5 Gene of Entamoeba histolytica Is Able to Direct a Transcription Start Site Independent of TATA or Initiator Regions". The Journal of Biological Chemistry 273 (34): 21663-8. doi:10.1074/jbc.273.34.21663. http://www.jbc.org/content/273/34/21663.full. Retrieved 2013-02-13.
- ↑ J. Carcamo, L. Buckbinder and D. Reinberg (1991). "The initiator directs the assembly of a transcription factor IID-dependent transcription complex". Proceedings of the National Academy of Sciences USA 88: 8052-6.
- ↑ L. Weis and D. Reinberg (1997). "Accurate positioning of RNA polymerase II on a natural TATA-less promoter is independent of TATA-binding protein associated factors and initiator-binding proteins". Mol. Cell. Biol. 17: 2973-84.
- 1 2 Tamar Juven-Gershon, Jer-Yuan Hsu, Joshua W. M. Theisen, and James T. Kadonaga (June 2008). "The RNA Polymerase II Core Promoter – the Gateway to Transcription". Current Opinion in Cell Biology 20 (3): 253-9. doi:10.1016/j.ceb.2008.03.003. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2586601/. Retrieved 2013-02-13.
- ↑ Noriyuki Sato; Tomohiro Katsuya; Hiromi Rakugi; Seiju Takami; Yukiko Nakata; Tetsuro Miki; Jitsuo Higaki; Toshio Ogihara (September 1997). "Association of Variants in Critical Core Promoter Element of Angiotensinogen Gene With Increased Risk of Essential Hypertension in Japanese". Hypertension 30 (3 Pt 1): 321-5. doi:10.1161/01.HYP.30.3.321. PMID 9314411. http://www.ncbi.nlm.nih.gov/pubmed/9314411. Retrieved 2012-02-20.
- 1 2 3 4 5 Dong-Hoon Lee, Naum Gershenzon, Malavika Gupta, Ilya P. Ioshikhes, Danny Reinberg and Brian A. Lewis (November 2005). "Functional Characterization of Core Promoter Elements: the Downstream Core Element Is Recognized by TAF1". Molecular and Cellular Biology 25 (21): 9674-86. doi:10.1128/MCB.25.21.9674-9686.2005. PMID 16227614. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1265815/. Retrieved 2010-10-23.
- ↑ Michael David Winther, Leah Christine Knickle, Martin Haardt, Stephen John Allen, Andre Ponton, Roberto Justo De Antueno, Kenneth Jenkins, Solomon O. Nwaka, Y. Paul Goldberg (July 29, 2004). "Fat Regulated Genes, Uses Thereof and Compounds for Mudulating Same". US Patent Office. Retrieved 2013-02-14.
- ↑ HGNC (February 5, 2013). "A1BG alpha-1-B glycoprotein [ Homo sapiens ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2013-02-14.
- 1 2 3 4 Thomas Abeel, Yves Van de Peer and Yvan Saeys (September 2009). "Toward a gold standard for promoter prediction evaluation". Bioinformatics 25 (12): i313-20. doi:10.1093/bioinformatics/btp191. http://bioinformatics.oxfordjournals.org/content/25/12/i313.full.pdf+html. Retrieved 2012-04-04.
- ↑ GA Patikoglou, JL Kim, L Sun, SH Yang, T Kodadek, SK Burley (1999). Genes Development 13: 3217-30.
- ↑ J. Wong, E. Bateman (1994). Nucleic Acids Research 22: 1890-96.
- ↑ JL Kim, DB Nikolov, SK Burley (1993). Nature 365: 520-7.
- ↑ YC Kim, JH Geiger, S Hahn, PB Sigler (1993). Nature 365: 512-20.
- ↑ J Corden, B Wasylyk, A Buchwalder, P Sassone-Corsi, C. Kedinger, P. Chambon (1980). Science 209: 1405-14.
- ↑ Cenik, C, et al. (2011). "Genome analysis reveals interplay between 5' UTR introns and nuclear mRNA export for secretory and mitochondrial genes". PLoS Genetics 7 (4). doi:10.1371/journal.pgen.1001366. http://www.plosgenetics.org/article/info:doi/10.1371/journal.pgen.1001366.
- ↑ "Five prime untranslated region, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 19, 2014. Retrieved 2014-05-16.
- ↑ Myer VE, Young RA (October 1998). "RNA polymerase II holoenzymes and subcomplexes". J. Biol. Chem. 273 (43): 27757–60. doi:10.1074/jbc.273.43.27757. PMID 9774381. http://www.jbc.org/cgi/reprint/273/43/27757.pdf.
- 1 2 "RNA polymerase II holoenzyme, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 1, 2014. Retrieved 2014-05-16.
- ↑ Kornberg R (1999). "Eukaryotic transcriptional control". Trends in Cell Biology 9 (12): M46. doi:10.1016/S0962-8924(99)01679-7. PMID 10611681.
- ↑ Klaus Hinkelmann, Oscar Kempthorne (2008). Design and Analysis of Experiments, Volume I: Introduction to Experimental Design (2nd ed.). Wiley. ISBN 978-0-471-72756-9. http://books.google.com/?id=T3wWj2kVYZgC&printsec=frontcover.
- ↑ R. A. Bailey (2008). Design of comparative experiments. Cambridge University Press. ISBN 978-0-521-68357-9. http://www.cambridge.org/uk/catalogue/catalogue.asp?isbn=9780521683579.
- ↑ "Treatment and control groups, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 18, 2012. Retrieved 2012-05-31.
- ↑ "proof of concept, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. November 10, 2012. Retrieved 2013-01-13.
- ↑ Ginger Lehrman and Ian B Hogue, Sarah Palmer, Cheryl Jennings, Celsa A Spina, Ann Wiegand, Alan L Landay, Robert W Coombs, Douglas D Richman, John W Mellors, John M Coffin, Ronald J Bosch, David M Margolis (August 13, 2005). "Depletion of latent HIV-1 infection in vivo: a proof-of-concept study". Lancet 366 (9485): 549-55. doi:10.1016/S0140-6736(05)67098-5. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1894952/. Retrieved 2012-05-09.
Further reading
- Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST (Jan 1994). "DNA Sequence Requirements for Transcriptional Initiator Activity in Mammalian Cells". Mol Cell Biol. 14 (1): 116-27. PMID 8264580. http://mcb.asm.org/cgi/reprint/14/1/116?view=long&pmid=8264580.
- Liston DR, Johnson PJ (Mar 1999). "Analysis of a Ubiquitous Promoter Element in a Primitive Eukaryote: Early Evolution of the Initiator Element". Mol Cell Biol. 19 (3): 2380-8. http://mcb.asm.org/cgi/content/full/19/3/2380.
- Myer VE, Young RA (October 1998). "RNA polymerase II holoenzymes and subcomplexes". J. Biol. Chem. 273 (43): 27757–60. doi:10.1074/jbc.273.43.27757. PMID 9774381. http://www.jbc.org/cgi/reprint/273/43/27757.pdf.
External links
- African Journals Online
- Bing Advanced search
- GenomeNet KEGG database
- Google Books
- Google scholar Advanced Scholar Search
- Home - Gene - NCBI
- JSTOR
- Lycos search
- NCBI All Databases Search
- NCBI Site Search
- PubChem Public Chemical Database
- Questia - The Online Library of Books and Journals
- SAGE journals online
- Scirus for scientific information only advanced search
- SpringerLink
- Taylor & Francis Online
- Virtual Cell Animation Collection, Introducing Transcription
- WikiDoc The Living Textbook of Medicine
- Wiley Online Library Advanced Search
- Yahoo Advanced Web Search
| |||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||
![]() This is a research project at http://en.wikiversity.org
This is a research project at http://en.wikiversity.org
| |
Development status: this resource is experimental in nature. |
| |
Educational level: this is a research resource. |
| |
Resource type: this resource is an article. |
| |
Resource type: this resource contains a lecture or lecture notes. |
| |
Subject classification: this is a biochemistry resource. |
| |
Subject classification: this is a genetics resource. |