RNA interference
The Nobel Prize in Physiology or Medicine in 2006 was awarded to Andrew Z. Fire and Craig C. Mello for their research on RNA interference [3]. The goal of this learning project is to complement the Wikipedia article about RNA interference in two ways. The first goal is to provide a user-friendly introduction to the topic. This means providing learning resources for people who would normally be unable to understand a technical Wikipedia article on the topic of RNA interference. The second goal is to provide learning resources that allow interested university students to collaboratively explore the science behind each awarded Nobel Prize in more detail than is possible with the related Wikipedia article. If you have not done so already, take a look at the Wikipedia article about RNA interference then select one of these learning paths:
- Explore a user-friendly introduction to the practical medical implications of RNA interference that arise from the Nobel Prize-winning scientific research of Andrew Z. Fire and Craig C. Mello.
- If you were able to read and appreciate the Wikipedia article about RNA interference then continue reading below and participate in further exploration of this subject.
RNA interference was recently discovered as a mechanism used by cells for regulating gene expression. This discovery has quickly resulted in the widespread use of artificial interfering RNAs as an important laboratory research technique for altering the amount of specific proteins inside cells. There is also active study of the potential value of RNA interference for medical applications[4].
Short review: RNA interference in cells
This section and the next few sections briefly introduce RNA interference (RNAi) and will orient you towards the activities. See the Wikipedia article about RNA interference for more a more detailed introduction to RNAi.
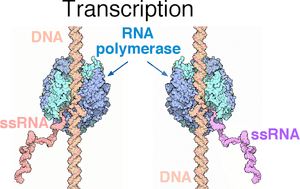
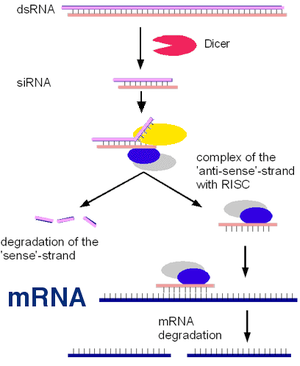
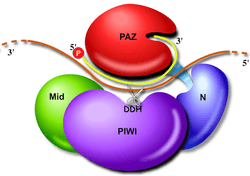
The normal function of RNA interference inside cells depends on the production of double stranded RNA (dsRNA). Base pair-complementary RNA strands (ssRNA) can be produced by transcription of both template DNA strands of some genes (Figure 1). In other cases, the protein-coding RNA sense strand might be produced by a virus and the antisense RNA strand produced by the host cell. The sense and antisense RNA strands form double strand RNA (Figure 2, top) that is processed to small (about 20 base pairs long) inhibitory RNA (siRNA). The siRNA can form a molecular complex with proteins that first strip away the sense strand of RNA, making the antisense inhibitory RNA (iRNA) available for base pairing with messenger RNA (mRNA). This targets a specific mRNA for destruction (Figure 3), resulting in inhibition of the biological function that would otherwise be served by the protein coded for by the mRNA.
There had long been interest in the idea that it might be possible to alter gene expression by using single strands of nucleic acid that might base pair hybridize with a specific target in cells. However, it was only in 1998 that experiments were described showing the unexpected power of double stranded RNA to block gene expression [6]. The experimental system used was the worm Caenorhabditis elegans. This was a useful experimental system because the developmental orgin of all of this organism's cells is known and it is possible to inject RNA into early embryos and observe changes in the pattern of development.
Protection against viruses
An important aspect of RNA interference is its role in protecting organisms from some deleterious effects of viruses [8]. Such a role for RNA interference was first found in plants, but has also been found in some animals[9].
Gene expression knockdown experiments
RNA interference is now a widely used biology research technique that can be applied to both cultured cells [10] and whole animals[11]. RNA interference can be used to selectively reduce the level of expression of a specific protein. Clues to the function of the protein can be obtained by observing changes in cell or organism behavior after knockdown of expression.
Medical applications
It has been suggested that knockdown of expression of proteins by virus-delivered siRNA might be effective for future treatments of Amyotrophic lateral sclerosis and other neurodegenerative diseases. Working with laboratory mice as an experimental model system for the human disease ALS, Miller et al showed that loss of muscle function could be slowed using RNA interference[12].
Learning project: what next?
Read about one of these topics and add what you learn to this page:
- What if inhibitory double stranded RNA could form from hairpins such as those found in tRNA molecules? (hint)
- What other medical applications have been proposed for RNA interference? (Hints: search here)
- Can dsRNAs also inhibit transcription? (Hint: start here)
- Add questions and general discussion to the next sections.
Your questions about RNAi
- ...
Can a ssRNA induce RNA interference?
Discussions
- Read RNAi: a novel strategy for the treatment of prion diseases by Qingzhong Kong and Lentivector-mediated RNAi efficiently suppresses prion protein and prolongs survival of scrapie-infected mice by Alexander Pfeifer et al. Discuss the idea that virus-mediated RNA interference can be used to treat neurological diseases.
Mice as a model system
Are short-lived mice a good model system for study of human neurodegenerative diseases?
References
- ↑ A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis by Manabu Yoshikawa, Angela Peragine, Mee Yeon Park and R. Scott Poethig in Genes Dev. (2005) Volume 19, pages 2164–2175.
- ↑ Discover the rules of DNA base pairing with an online simulator. The base pairing rules for RNA are similar.
- ↑ 2006 award at the Nobel Prize website.
- ↑ RNA interference by Julian Downward in British Medical Journal (2004) Volume 328: pages 1245–1248.
- ↑ An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells by S. M. Hammond, E. Bernstein, D. Beach and G. J. Hannon in Nature (2000) Volume 404, pages 293-296.
- ↑ Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans by A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello in Nature (1998) Volume 391, pages 806-811.
- ↑ "Purified Argonaute2 and an siRNA form recombinant human RISC" by F. V. Rivas, N. H. Tolia, J. J. Song, J. P. Aragon, J. Liu, G. J. Hannon and L. Joshua-Tor in Nature structural & molecular biology (2005) Volume 12, pages 340-349.
- ↑ Review for the 2006 Nobel Prize in Physiology or Medicine, Advanced Information: RNA interference by Bertil Daneholt.
- ↑ Antiviral silencing in animals by Hong-Wei Li and Shou-Wei Ding in FEBS Lett. (2005) Volume 579, pages 5965–5973.
- ↑ Transient and Stable Knockdown of the Integrase Cofactor LEDGF/p75 Reveals Its Role in the Replication Cycle of Human Immunodeficiency Virus by L. Vandekerckhove, F. Christ, B. Van Maele, J. De Rijck, R. Gijsbers, C. Van den Haute, M. Witvrouw and Z. Debyser in Journal of Virology (2006) Volume 80, pages 1886-1896.
- ↑ Heritable and stable gene knockdown in rats by C. T. Dann, A. L. Alvarado, R. E. Hammer and D. L. Garbers in Proceedings of the National Academy of Sciences U.S.A. (2006) Volume 103, pages 11246-11251.
- ↑ Virus-Delivered Small RNA Silencing Sustains Strength in Amyotrophic Lateral Sclerosis by T. M. Miller, B. K. Kaspar, G. J. Kops, K. Yamanaka, L. J. Christian, F. H. Gage and D. W. Cleveland in Annals of neurology (2006) Volume 57, pages 773-776.
See also
- This lesson on RNA interference is part of the Nobel Prize in Physiology or Medicine learning project.
- Nobel Prize in Chemistry learning project
- RNA World
- Cell biology improvement drive
External links
- RNA interference article at Wikipedia.