Gene transcriptions
DNA is a double helix of interlinked nucleotides surrounded by an epigenome. On the basis of biochemical signals, an enzyme, specifically a ribonucleic acid (RNA) polymerase, is chemically bonded to one of the strands (the template strand) of this double helix. The polymerase, once phosphorylated, begins to catalyze the formation of RNA using the template strand. Although the catalysis may have more than one beginning nucleotide (a start site) and more than one ending nucleotide (a stop site) along the DNA, each nucleotide sequence catalyzed that ultimately produces approximately the same RNA is part of a gene. The catalysis of each RNA representation from the template DNA is a transcription, specifically a gene transcription. The overall process is also referred to as gene transcription.
Deoxyribonucleic acid
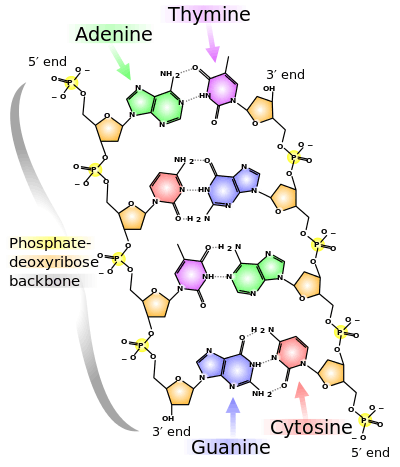
Deoxyribonucleic acid (DNA) is a polymer composed of nucleic acids linked together with the sugar deoxyribose.
Genes
Def. "[a] unit of heredity; a segment of DNA or RNA that is transmitted from one generation to the next, and that carries genetic information such as the sequence of amino acids for a protein"[1] is called a gene.
Promoters
Def. "[t]he section of DNA that controls the initiation of RNA transcription as a product of a gene"[2] is called a gene promoter, or a promoter in the field of genetics.
Proximal promoters
Def. any proximal nucleotide "sequence upstream of the gene that tends to contain primary regulatory elements"[3] is called a proximal promoter.
Core promoters
"The core promoter is the minimal portion of the promoter required to properly initiate gene transcription.[4]"[3] It contains a binding site for RNA polymerase (RNA polymerase I, RNA polymerase II, or RNA polymerase III).
The core promoter is approximately -34 nt upstream from the TSS.
Senses
"[A] single strand of DNA [has a positive sense (+)] if an RNA version of the same sequence is translated or translatable into protein. Its complementary strand is called antisense (or negative (-) sense). Sometimes the phrase coding strand is encountered; however, protein coding and non-coding RNA's can be transcribed similarly from both strands, in some cases being transcribed in both directions from a common promoter region, or being transcribed from within introns, on both strands".[5][6][7]"[8]
"The two complementary strands of double-stranded DNA (dsDNA) are usually differentiated as the "sense" strand and the "antisense" strand. The DNA sense strand looks like the messenger RNA (mRNA) and can be used to read the expected protein code by human eyes (e.g. ATG codon = Methionine amino acid). However, the DNA sense strand itself is not used to make protein by the cell. It is the DNA antisense strand which serves as the source for the protein code, because, with bases complementary to the DNA sense strand, it is used as a template for the mRNA. Since transcription results in an RNA product complementary to the DNA template strand, the mRNA is complementary to the DNA antisense strand. The mRNA is what is used for translation (protein synthesis)."[8]
"The only real biological information that is important for labeling strands is the location of the 5' phosphate group and the 3' hydroxyl group because these ends determine the direction of transcription and translation. A sequence 5' CGCTAT 3' is equivalent to a sequence written 3' TATCGC 5' as long as the 5' and 3' ends are noted. If the ends are not labeled, convention is to assume that the sequence is written in the 5' to 3' direction. Good rule of thumb for figuring out the "sense" strand: Look for the start codon ATG (AUG in mRNA). In the table example, the sense mRNA has the AUG codon at the end (remember that translation proceeds in the 5' to 3' direction)."[8]
Preinitiation complexes

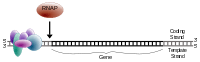
For eukaryotic transcription, the RNA polymerase II holoenzyme de-helicizes the DNA, attaches along the template strand.
Once the preinitiation complex has found its appropriate attachment section along the template strand of DNA, RNA polymerase II is attached and begins transcription.
RNA polymerase II holoenzyme complexes
"RNA polymerase II ... is recruited to the promoters of protein-coding genes in living cells.[9]"[10] Or, transcription factories are present and the euchromatin is brought within the nearest transcription factory and A1BG messenger RNA (mRNA) is transcribed.
For those circumstances in which the holoenzyme is built onto the euchromatin, it is necessary to consider the holoenzyme components and the likely sequence of binding, RNA polymerase II entrance upon the scene and subsequent action.
"RNA polymerase II (also called RNAP II and Pol II) ... catalyzes the transcription of DNA to synthesize precursors of mRNA and most snRNA and microRNA.[11] ... In humans RNAP II consists of seventeen protein molecules (gene products encoded by POLR2A-L, where the proteins synthesized from 2C-, E-, and F-form homodimers)."[10]
RNA polymerase II holoenzyme complex may also have to search for one or more transcription start sites.
Transcription start sites
The transcription start site "is the location where transcription starts at the 5'-end of a gene sequence.[12]"[13]
A start site is a biochemically signaled nucleotide or set of nucleotides for attachment either to the epigenome or the DNA.
Research
Hypothesis:
- Gene transcription can occur for each gene on either strand (template or coding).
- Gene transcription can occur for each gene in either direction (+ or -).
Control groups

The findings demonstrate a statistically systematic change from the status quo or the control group.
“In the design of experiments, treatments [or special properties or characteristics] are applied to [or observed in] experimental units in the treatment group(s).[14] In comparative experiments, members of the complementary group, the control group, receive either no treatment or a standard treatment.[15]"[16]
Proof of concept
Def. a “short and/or incomplete realization of a certain method or idea to demonstrate its feasibility"[17] is called a proof of concept.
Def. evidence that demonstrates that a concept is possible is called proof of concept.
The proof-of-concept structure consists of
- background,
- procedures,
- findings, and
- interpretation.[18]
See also
References
- ↑ "gene, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. December 13, 2012. Retrieved 2012-12-13.
- ↑ "promoter, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. September 20, 2012. Retrieved 2012-09-29.
- 1 2 "Promoter (genetics), In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. September 16, 2012. Retrieved 2012-09-29.
- ↑ Stephen T. Smale and James T. Kadonaga (July 2003). "The RNA Polymerase II Core Promoter". Annual Review of Biochemistry 72 (1): 449-79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. http://www.annualreviews.org/doi/abs/10.1146/annurev.biochem.72.121801.161520. Retrieved 2012-05-07.
- ↑ Anne-Lise Haenni (2003). "Expression strategies of ambisense viruses". Virus Research 93 (2): 141–150. doi:10.1016/S0168-1702(03)00094-7. PMID 12782362. http://www.sciencedirect.com/science/article/pii/S0168170203000947.
- ↑ Kakutani T, Hayano Y, Hayashi T, Minobe Y (1991). "Ambisense segment 3 of rice stripe virus: the first instance of a virus containing two ambisense segments". J Gen Virol. 72: 465–8. PMID 1993885.
- ↑ Zhu Y, Hayakawa T, Toriyama S, Takahashi M (1991). "Complete nucleotide sequence of RNA 3 of rice stripe virus: an ambisense coding strategy". J Gen Virol 72: 763–7. PMID 2016591.
- 1 2 3 "Sense (molecular biology), In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. February 12, 2013. Retrieved 2013-02-14.
- ↑ Myer VE, Young RA (October 1998). "RNA polymerase II holoenzymes and subcomplexes". J. Biol. Chem. 273 (43): 27757–60. doi:10.1074/jbc.273.43.27757. PMID 9774381. http://www.jbc.org/cgi/reprint/273/43/27757.pdf.
- 1 2 "RNA polymerase II holoenzyme, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 1, 2014. Retrieved 2014-05-16.
- ↑ Kornberg R (1999). "Eukaryotic transcriptional control". Trends in Cell Biology 9 (12): M46. doi:10.1016/S0962-8924(99)01679-7. PMID 10611681.
- ↑ Marketa Zvelebil, Jeremy O. Baum (2008). Understanding bioinformatics. Garland Science. ISBN 978-0815340249.
- ↑ Ino4president (November 4, 2011). "Transcription start site, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. Retrieved 2012-05-16.
- ↑ Klaus Hinkelmann, Oscar Kempthorne (2008). Design and Analysis of Experiments, Volume I: Introduction to Experimental Design (2nd ed.). Wiley. ISBN 978-0-471-72756-9. http://books.google.com/?id=T3wWj2kVYZgC&printsec=frontcover.
- ↑ R. A. Bailey (2008). Design of comparative experiments. Cambridge University Press. ISBN 978-0-521-68357-9. http://www.cambridge.org/uk/catalogue/catalogue.asp?isbn=9780521683579.
- ↑ "Treatment and control groups, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 18, 2012. Retrieved 2012-05-31.
- ↑ "proof of concept, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. November 10, 2012. Retrieved 2013-01-13.
- ↑ Ginger Lehrman and Ian B Hogue, Sarah Palmer, Cheryl Jennings, Celsa A Spina, Ann Wiegand, Alan L Landay, Robert W Coombs, Douglas D Richman, John W Mellors, John M Coffin, Ronald J Bosch, David M Margolis (August 13, 2005). "Depletion of latent HIV-1 infection in vivo: a proof-of-concept study". Lancet 366 (9485): 549-55. doi:10.1016/S0140-6736(05)67098-5. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1894952/. Retrieved 2012-05-09.
External links
- African Journals Online
- Bing Advanced search
- GenomeNet KEGG database
- Google Books
- Google scholar Advanced Scholar Search
- Home - Gene - NCBI
- JSTOR
- Lycos search
- NCBI All Databases Search
- NCBI Site Search
- Office of Scientific & Technical Information
- PubChem Public Chemical Database
- Questia - The Online Library of Books and Journals
- SAGE journals online
- Scirus for scientific information only advanced search
- SpringerLink
- Taylor & Francis Online
- WikiDoc The Living Textbook of Medicine
- Wiley Online Library Advanced Search
- Yahoo Advanced Web Search
| |||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||
![]() This is a research project at http://en.wikiversity.org
This is a research project at http://en.wikiversity.org
| |
Development status: this resource is experimental in nature. |
| |
Educational level: this is a research resource. |
| |
Resource type: this resource is an article. |
| |
Resource type: this resource contains a lecture or lecture notes. |
| |
Subject classification: this is a biochemistry resource. |
| |
Subject classification: this is a genetics resource. |
| |
Subject classification: this is a medicine resource. |