Epigenomes
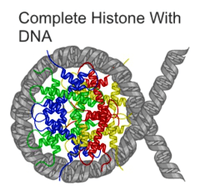
Inside each eukaryote nucelus is genetic material (DNA) surrounded by protective and regulatory proteins. These protective and regulatory proteins and the dynamic changes to them that occur during the course of a eukaryote's existence are the epigenome.
Epigenomic theory
Def. a chemical entity anterior to, after, at, besides, near to, on, outer to, over, related to, or upon another chemical is called an epi (or epi-) chemical.
Genomes
"[T]he genome is the entirety of an organism's hereditary information. [In humans, it] is encoded ... in DNA ... The genome includes both the genes and the non-coding sequences of the DNA.[1]"[2]
"Homo sapiens estimated genome size [is] 3.2 billion bp.[3]"[2]
Genetic information is encoded as a sequence of nucleobases: adenine (A), cytosine (C), guanine (G), and thymine (T).
Deoxyribonucleic acid molecules
Deoxyribonucleic acid (DNA) is composed of nucleobases (the sequence of which is the genome), deoxyribose (a sugar), and phosphate groups. Each nucleobase is attached to one deoxyribose molecule and one (PO4) phosphate molecule to form a chain of nucleotides (nucleobase + deoxyribose + phosphate) for a haploid genome. A linking of nucleobases may occur without the phosphate or the deoxyribose. The phosphate and the sugar are part of the epigenome.
DNA often occurs as a double helix. The linking between one nitrogenous nucleobase of a DNA molecule and another nitrogenous nucleobase of a second DNA molecule is via hydrogen bonds. Each hydrogen bond ("the electromagnetic attractive interaction of a hydrogen atom and an electronegative atom, such as nitrogen [or] oxygen"[4] of a nucleobase) is part of the epigenome.
Nucleosomes
DNA packaging in eukaryotes consists of "DNA wound in sequence around four histone protein cores.[5]"[6]
"Nucleosomes form the fundamental repeating units of eukaryotic chromatin[7]".[6]
"The nucleosome core particle consists of approximately 147 base pairs of DNA wrapped in 1.67 left-handed superhelical turns around a histone octamer consisting of 2 copies each of the core histones H2A, H2B, H3, and H4.[8]"[6]
"Core particles are connected by stretches of "linker DNA", which can be up to about 80 bp long."[6]
Histones
"Histone deacetylases (HDAC) ([Enzyme Commission number] EC number 3.5.1) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on a histone, allowing the histones to wrap the DNA more tightly."[9]
"[Histone deacetylase] action is opposite to that of histone acetyltransferase."[9]
Euchromatin
Def. "uncoiled dispersed threads of chromosomal material that occurs during interphase"[10] is called euchromatin.
"The structure of euchromatin is reminiscent of an unfolded set of beads along a string, wherein those beads represent nucleosomes."[11]
"[T]he presence of methylated lysine 4 on the histone tails [may act] as a general marker for euchromatin."[11]
"One example of constitutive euchromatin that is 'always turned on' is housekeeping genes, which code for the proteins needed for basic functions of cell survival."[11]
Heterochromatin
"Heterochromatin mainly consists of genetically inactive satellite sequences,[12] and many genes are repressed to various extents, although some cannot be expressed in euchromatin at all.[13] Both centromeres and telomeres are heterochromatic, as is the Barr body of the second, inactivated X-chromosome in a female."[14]
Constitutive heterochromatin
"[S]ections of DNA that occur ... particularly at the centromeres and telomeres ... often consisting of ... repetitive DNA [that is largely] transcriptionally silent" are constitutive heterochromatin.[15]
"[R]egions of DNA that exist as constitutive heterochromatin [are] the same for all cells of a given species."[15]
"[A]ll human chromosomes 1, 9, 16, and the Y-chromosome contain large regions of constitutive heterochromatin. In most organisms, constitutive heterochromatin occurs around the chromosome centromere and near telomeres."[14]
Facultative heterochromatin
"[G]enes that are silenced through a mechanism such as histone methylation or siRNA through RNAi [produce facultative heterochromatin]."[14]
"The regions of DNA packaged in facultative heterochromatin [are not] consistent between the cell types within a species, and thus a sequence in one cell that is packaged in facultative heterochromatin (and the genes within poorly expressed) may be packaged in euchromatin in another cell (and the genes within no longer silenced)."[14]
"An example of facultative heterochromatin is X-chromosome inactivation in female mammals: one X chromosome is packaged as facultative heterochromatin and silenced, while the other X chromosome is packaged as euchromatin and expressed."[14]
Changes
“Changes to the epigenome can result in changes to the structure of chromatin and changes to the function of the genome.[16]”[17]
There are "nearly 50,000 acetylated sites [punctate sites of modified histones] in the human genome that correlate with active transcription start sites and CpG islands and tend to cluster within gene-rich loci."[16]
Acetyl groups
"Acetylation (or in IUPAC nomenclature ethanoylation) describes a reaction that introduces an acetyl functional group into a chemical compound. (Deacetylation is the removal of the acetyl group.)"[18]
"In histone acetylation and deacetylation, histone proteins are acetylated and deacetylated on lysine residues in the N-terminal tail as part of gene regulation. Typically, these reactions are catalyzed by enzymes with histone acetyltransferase (HAT) or histone deacetylase (HDAC) activity, although HATs and HDACs can modify the acetylation status of non-histone proteins as well.[19]"[18]
"[L]ysine acetylation almost always correlates with chromatin accessibility and transcriptional activity"[16].
Methyl groups
Methylation is "the addition of a methyl group ... replacing a hydrogen atom."[20]
"DNA methylation in vertebrates typically occurs at CpG sites (cytosine-phosphate-guanine sites, that is, where a cytosine is directly followed by a guanine in the DNA sequence). This methylation results in the conversion of the cytosine to 5-methylcytosine. The formation of Me-CpG is catalyzed by the enzyme DNA methyltransferase. Human DNA has about 80%-90% of CpG sites methylated, but there are certain areas, known as CpG islands, that are GC-rich (made up of about 65% CG residues), wherein none are methylated. These are associated with the promoters of 56% of mammalian genes, including all ubiquitously expressed genes. One to two percent of the human genome are CpG clusters, and there is an inverse relationship between CpG methylation and transcriptional activity."[20]
"Non-CpG methylation (CNG and CNN) ... has been observed at a low frequency in the early mouse embryo"[16]
"Protein methylation typically takes place on arginine or lysine amino acid residues in the protein sequence.[21] Arginine can be methylated once (monomethylated arginine) or twice, with either both methyl groups on one terminal nitrogen (asymmetric dimethylated arginine) or one on both nitrogens (symmetric dimethylated arginine) by peptidylarginine methyltransferases (PRMTs). Lysine can be methylated once, twice or three times by lysine methyltransferases. Protein methylation has been most-studied in the histones. The transfer of methyl groups from S-adenosyl methionine to histones is catalyzed by enzymes known as histone methyltransferases. Histones that are methylated on certain residues can act epigenetically to repress or activate gene expression.[22][23]"[20]
Phosphoryl groups
"Phosphorylation is the addition of a phosphate (PO43-) group to a protein or other organic molecule."[24]
"Kinases phosphorylate proteins and phosphatases dephosphorylate proteins."[24]
"Reversible phosphorylation of proteins is an important regulatory mechanism that occurs in both prokaryotic and eukaryotic organisms.[25][26][27][28]"[24]
Phosphoryl groups attach to histones at serine and threonine sites.[16]
Ubiquityl groups
"The core histones that make up the nucleosome are subject to ... modifications, including ubiquitination [that occurs] primarily at specific positions within the amino-terminal histone tails."[16]
Research
Hypothesis:
- The epigenome around A1BG is opened as if for any gene rather than a specific promoter, enhancer, or other transcription related factor.
Control groups

The findings demonstrate a statistically systematic change from the status quo or the control group.
“In the design of experiments, treatments [or special properties or characteristics] are applied to [or observed in] experimental units in the treatment group(s).[29] In comparative experiments, members of the complementary group, the control group, receive either no treatment or a standard treatment.[30]"[31]
Proof of concept
Def. a “short and/or incomplete realization of a certain method or idea to demonstrate its feasibility"[32] is called a proof of concept.
Def. evidence that demonstrates that a concept is possible is called proof of concept.
The proof-of-concept structure consists of
- background,
- procedures,
- findings, and
- interpretation.[33]
See also
References
- ↑ Ridley, M. (2006). Genome. New York, NY: Harper Perennial. ISBN 0-06-019497-9
- 1 2 "Genome, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. October 19, 2012. Retrieved 2012-10-30.
- ↑ "Human Genome".
- ↑ "Hydrogen bond, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. October 27, 2012. Retrieved 2012-10-30.
- ↑ Reece, Jane; Campbell, Neil (2006). Biology. San Francisco: Benjamin Cummings. ISBN 0-8053-6624-5.
- 1 2 3 4 "Nucleosome, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. October 17, 2012. Retrieved 2012-10-29.
- ↑ Alberts, Bruce (2002). Molecular biology of the cell (4th ed.). New York: Garland Science. p. 207. ISBN 0-8153-4072-9. http://www.ncbi.nlm.nih.gov/books/bv.fcgi?highlight=Nucleosome&rid=mboc4.section.608#630.
- ↑ Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (September 1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution". Nature 389 (6648): 251–60. doi:10.1038/38444. PMID 9305837.
- 1 2 "Histone deacetylase, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. October 26, 2012. Retrieved 2012-10-29.
- ↑ "euchromatin, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. January 2, 2012. Retrieved 2012-10-30.
- 1 2 3 "Euchromatin, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. October 20, 2012. Retrieved 2012-10-30.
- ↑ Lohe, A.R., et al. (August 1, 1993). "Mapping Simple Repeated DNA Sequences in Heterochromatin of Drosophila Melanogaster". Genetics 134 (4): 1149–74. ISSN 0016-6731. PMID 8375654. PMC 1205583. http://www.genetics.org/cgi/content/full/134/4/1149.
- ↑ Lu, B.Y., et al. (June 1, 2000). "Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila". Genetics 155 (2): 699–708. ISSN 0016-6731. PMID 10835392. PMC 1461102. http://www.genetics.org/cgi/content/full/155/2/699.
- 1 2 3 4 5 "Heterochromatin, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. October 29, 2012. Retrieved 2012-10-29.
- 1 2 "Constitutive heterochromatin, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. September 28, 2012. Retrieved 2012-10-29.
- 1 2 3 4 5 6 Bradley E. Bernstein, Alexander Meissner, Eric S. Lander (February 23, 2007). "The Mammalian Epigenome". Cell 128 (4): 669–81. doi:10.1016/j.cell.2007.01.033. http://www.sciencedirect.com/science/article/pii/S0092867407001286. Retrieved 19 December 2011.
- ↑ "Epigenome, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. September 17, 2012. Retrieved 2012-10-29.
- 1 2 "Acetylation, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. September 4, 2012. Retrieved 2012-10-30.
- ↑ Sadoul K, Boyault C, Pabion M, Khochbin S (February 2008). "Regulation of protein turnover by acetyltransferases and deacetylases". Biochimie 90 (2): 306–12. doi:10.1016/j.biochi.2007.06.009. PMID 17681659.
- 1 2 3 "Methylation, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. August 20, 2012. Retrieved 2012-10-30.
- ↑ Christopher Walsh (2006). "Chapter 5 - Protein Methylation". Posttranslational modification of proteins: expanding nature's inventory. Roberts and Co. Publishers. ISBN 0-9747077-3-2. http://www.roberts-publishers.com/walsh/chapter5.pdf.
- ↑ Grewal SI, Rice JC (2004). "Regulation of heterochromatin by histone methylation and small RNAs". Curr. Opin. Cell Biol. 16 (3): 230–8. doi:10.1016/j.ceb.2004.04.002. PMID 15145346. http://linkinghub.elsevier.com/retrieve/pii/S0955067404000535.
- ↑ Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001). "Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly". Science 292 (5514): 110–3. doi:10.1126/science.1060118. PMID 11283354. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=11283354.
- 1 2 3 "Phosphorylation, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. October 29, 2012. Retrieved 2012-10-30.
- ↑ Cozzone AJ (1988). "Protein phosphorylation in prokaryotes". Annu. Rev. Microbiol. 42: 97–125. doi:10.1146/annurev.mi.42.100188.000525. PMID 2849375.
- ↑ Stock JB, Ninfa AJ, Stock AM (December 1989). "Protein phosphorylation and regulation of adaptive responses in bacteria". Microbiol. Rev. 53 (4): 450–90. PMID 2556636. PMC 372749. //www.ncbi.nlm.nih.gov/pmc/articles/PMC372749/.
- ↑ Chang C, Stewart RC (July 1998). "The Two-Component System . Regulation of Diverse Signaling Pathways in Prokaryotes and Eukaryotes". Plant Physiol. 117 (3): 723–31. doi:10.1104/PPSOE.117.3.723. PMID 9662515. PMC 1539182. //www.ncbi.nlm.nih.gov/pmc/articles/PMC1539182/.
- ↑ Barford D, Das AK, Egloff MP (1998). "The structure and mechanism of protein phosphatases: insights into catalysis and regulation". Annu Rev Biophys Biomol Struct 27: 133–64. doi:10.1146/annurev.biophys.27.1.133. PMID 9646865.
- ↑ Klaus Hinkelmann, Oscar Kempthorne (2008). Design and Analysis of Experiments, Volume I: Introduction to Experimental Design (2nd ed.). Wiley. ISBN 978-0-471-72756-9. http://books.google.com/?id=T3wWj2kVYZgC&printsec=frontcover.
- ↑ R. A. Bailey (2008). Design of comparative experiments. Cambridge University Press. ISBN 978-0-521-68357-9. http://www.cambridge.org/uk/catalogue/catalogue.asp?isbn=9780521683579.
- ↑ "Treatment and control groups, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 18, 2012. Retrieved 2012-05-31.
- ↑ "proof of concept, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. November 10, 2012. Retrieved 2013-01-13.
- ↑ Ginger Lehrman and Ian B Hogue, Sarah Palmer, Cheryl Jennings, Celsa A Spina, Ann Wiegand, Alan L Landay, Robert W Coombs, Douglas D Richman, John W Mellors, John M Coffin, Ronald J Bosch, David M Margolis (August 13, 2005). "Depletion of latent HIV-1 infection in vivo: a proof-of-concept study". Lancet 366 (9485): 549-55. doi:10.1016/S0140-6736(05)67098-5. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1894952/. Retrieved 2012-05-09.
External links
- African Journals Online
- Bing Advanced search
- GenomeNet KEGG database
- Google Books
- Google scholar Advanced Scholar Search
- Home - Gene - NCBI
- JSTOR
- Lycos search
- NCBI All Databases Search
- Office of Scientific & Technical Information
- PsycNET
- PubChem Public Chemical Database
- Questia - The Online Library of Books and Journals
- SAGE journals online
- Scirus for scientific information only advanced search
- SpringerLink
- Taylor & Francis Online
- WikiDoc The Living Textbook of Medicine
- Wiley Online Library Advanced Search
- Yahoo Advanced Web Search
| |||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||
![]() This is a research project at http://en.wikiversity.org
This is a research project at http://en.wikiversity.org
| |
Development status: this resource is experimental in nature. |
| |
Educational level: this is a research resource. |
| |
Resource type: this resource is an article. |
| |
Resource type: this resource contains a lecture or lecture notes. |
| |
Subject classification: this is a biochemistry resource. |
| |
Subject classification: this is a medicine resource. |