Deoxyribonucleic acids
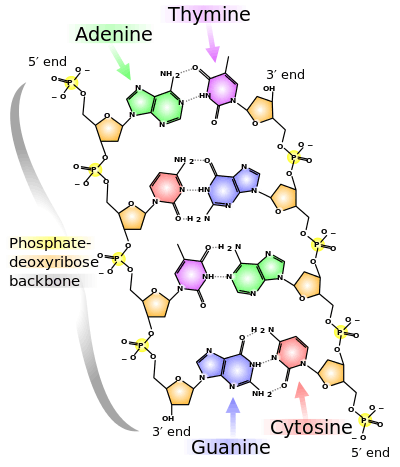
Deoxyribonucleic acid (DNA) is a polymer composed of nucleic acids linked together by a sugar-phosphate backbone.
The nucleic acids are inorganic acids with phosphoric acid as the only acid.
Attached to each sugar is a nucleobase.
Polyphosphoric acids
Def. "[a] colourless liquid, H3PO4"[1] is called phosphoric acid, orthophosphoric acid, or monophosphoric acid.
"[A]n orthophosphoric acid molecule can dissociate up to three times, giving up an H+ each time, which typically combines with a water molecule, H2O, as shown in these [chemical] reactions:"[2]
- H3PO4(s) + H2O(l) ↔ H3O+(aq) + H2PO4−(aq) Ka1= 7.25×10−3
- H2PO4−(aq)+ H2O(l) ↔ H3O+(aq) + HPO42−(aq) Ka2= 6.31×10−8
- HPO42−(aq)+ H2O(l) ↔ H3O+(aq) + PO43−(aq) Ka3= 4.80×10−13
"The anion after the first dissociation, H2PO4−, is the dihydrogen phosphate anion. The anion after the second dissociation, HPO42−, is the hydrogen phosphate anion. The anion after the third dissociation, PO43−, is the phosphate or orthophosphate anion. For each of the dissociation reactions shown above, there is a separate acid dissociation constant, called Ka1, Ka2, and Ka3 given at 25 °C. Associated with these three dissociation constants are corresponding pKa1=2.12, pKa2=7.21, and pKa3=12.67 values at 25 °C. Even though all three hydrogen (H) atoms are equivalent on an orthophosphoric acid molecule, the successive Ka values differ since it is energetically less favorable to lose another H+ if one (or more) has already been lost and the molecule/ion is more negatively charged."[2]
"For a given total acid concentration [A] = [H3PO4] + [H2PO4−] + [HPO42−] + [PO43−] ([A] is the total number of moles of pure H3PO4 which have been used to prepare 1 liter of solution), the composition of an aqueous solution of phosphoric acid can be calculated using the equilibrium equations associated with the three reactions described above together with the [H+][OH−] = 10−14 relation and the electrical neutrality equation. Possible concentrations of polyphosphoric molecules and ions is neglected. The system may be reduced to a fifth degree equation for [H+] which can be solved numerically, yielding:"[2]
| [A] (mol/L) | pH | [H3PO4]/[A] (%) | [H2PO4−]/[A] (%) | [HPO42−]/[A] (%) | [PO43−]/[A] (%) |
|---|---|---|---|---|---|
| 1 | 1.08 | 91.7 | 8.29 | 6.20×10−6 | 1.60×10−17 |
| 10−1 | 1.62 | 76.1 | 23.9 | 6.20×10−5 | 5.55×10−16 |
| 10−2 | 2.25 | 43.1 | 56.9 | 6.20×10−4 | 2.33×10−14 |
| 10−3 | 3.05 | 10.6 | 89.3 | 6.20×10−3 | 1.48×10−12 |
| 10−4 | 4.01 | 1.30 | 98.6 | 6.19×10−2 | 1.34×10−10 |
| 10−5 | 5.00 | 0.133 | 99.3 | 0.612 | 1.30×10−8 |
| 10−6 | 5.97 | 1.34×10−2 | 94.5 | 5.50 | 1.11×10−6 |
| 10−7 | 6.74 | 1.80×10−3 | 74.5 | 25.5 | 3.02×10−5 |
| 10−10 | 7.00 | 8.24×10−4 | 61.7 | 38.3 | 8.18×10−5 |
"For large acid concentrations, the solution is mainly composed of H3PO4. For [A] = 10−2, the pH is close to pKa1, giving an equimolar mixture of H3PO4 and H2PO4−. For [A] below 10−3, the solution is mainly composed of H2PO4− with [HPO42−] becoming non negligible for very dilute solutions. [PO43−] is always negligible. Note that the above analysis does not take into account ion activity coefficients; as such, the pH and molarity of a real phosphoric acid solution may deviate substantially from the above values."[2]
Def. "any of a class of inorganic polymers containing linked phosphate groups"[3], or more than five linked phosphoric acids, is called a polyphosphate, or polyphosphoric acid.
Def. typically two to twenty, or three to seven, linked monophosphoric acids, or orthophosphoric acids, is called an oligophosphoric acid.
Sugars
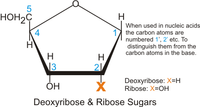
Def. "[a] derivative of the pentose sugar ribose in which the 2' hydroxyl (-OH) is reduced to a hydrogen (H)"[4] is called deoxyribose.
In the diagram of DNA at the page top, the pentose sugar deoxyribose is in a cyclic, furanose (5-membered ring) form. D-deoxyribose or L-deoxyribose is not demonstrated, but is determined by the hydroxyls being primarily below (D) or above (L) the plane of the ring. In Earth-based DNA, deoxyribose is in the dextro (D) configuration.
Deoxyribose may also occur in a pyranose (six-membered ring) form.
There are other pentose sugars including aldopentoses: apiose, arabinose, xylose, and lyxose, and ketopentoses: ribulose and xylulose. These may have a deoxy-form: deoxyapiose, deoxyarabinose, deoxyxylose, deoxylyxose, deoxyribulose, and deoxyxylulose. They may occur as a levo or dextro sugar and as a furanose or pyranose.
To occur in an Earth-like DNA, each of these six deoxypentoses and perhaps other sugars need to be dextro furanoses. Each is a DNA, for example, deoxyapionucleic acid.
Nitrogenous bases
Nitrogenous bases, found in cell nuclei, are nucleobases.
"In normal spiral DNA the bases form pairs between the two strands: [Adenine] A with [Thymine] T and [Cytosine] C with [Guanine] G. Purines pair with pyrimidines mainly for dimensional reasons - only this combination fits the constant width geometry of the DNA spiral."[5]
Nitrogenous bases include purines: adenine, guanine, hypoxanthine, isoguanine, xanthine, and 7-methylguanine; and pyrimidines: cytosine, thymine, and isocytosine.
Nucleic acids

"Synthetic genetics is a subdiscipline of synthetic biology that aims to develop artificial genetic polymers (also referred to as xeno-nucleic acids or XNAs) that can replicate in vitro and eventually in model cellular organisms."[6]
Def. any "acidic, chainlike biological macromolecule consisting of multiply repeat units of phosphoric acid, sugar and purine and pyrimidine bases"[7] occurring in cell nuclei is called a nucleic acid.
Def. a nucleic acid "in which the sugar component is threose"[8] is called threose nucleic acid, or threonucleic acid (TNA).
Additional DNAs may be
- deoxyapionucleic acid,
- deoxyarabinonucleic acid,
- deoxyxylonucleic acid (dXyNA),
- deoxylyxonucleic acid,
- deoxyribulonucleic acid, and
- deoxyxylulonucleic acid.
Synthesis of deoxyapionucleic acid has been accomplished.[9]
Deoxyxylonucleic acid and xylose nucleic acid have been produced.[10]
"[X]ylonucleic acid (XyloNA) [contains] a potentially prebiotic xylose sugar (a 3′-epimer of ribose) in its backbone."[10]
A "number of sugar-modified nucleic acid variants has been revealed as new genetic polymers, (2) some of them are endowed with catalytic activity (for e.g. FANA and HNA) (3). The structure of these artificial nucleic acids, however, mimics natural nucleic acid helicity (4)."[10]
"Although helices display a distinct pitch and curvature, they feature ca. 11–12 base pairs per turn, and χ/δ covariance plots indicate that the backbones of XNA:RNA or XNA:DNA heteroduplexes adopt an architecture that is either closely related to the A-form, as in the case of [1,5-anhydrohexitol nucleic acid (HNA)] HNA:RNA (96), [locked nucleic acid (LNA)] LNA:RNA (83), [cyclohexene nucleic acid (CeNA)] CeNA:RNA (85) and PNA:RNA (59), or between the A- and B-forms, as seen in the structures of DNA:RNA (97), [arabinonucleic acid (ANA)] ANA:RNA (79), [2′-deoxy-2′-fluoro-arabinonucleic acid (FANA)]FANA:RNA (79) and [peptide nucleic acid (PNA)] PNA:DNA (98)."[6]
Additional XNAs include bridged nucleic acid (BNA) glycol nucleic acid (GNA), FANA and peptide nucleic acid (PNA).
On the right is a diagram displaying various artificial and natural nucleic acid polymers.
"Representative structures illustrate the structural diversity and plasticity of natural and artificial nucleic acid (XNA) backbones. Structures are shown in alphabetic order. (A) Natural genetic polymers: B-form DNA (black), DNA:RNA hybrid and A-form RNA (gray). (B) Representative structures of XNA heteroduplexes with RNA or DNA. The RNA strand is shown in gray, the DNA strand in black and the orientation of the XNA strand is indicated. (C) XNA homoduplexes. Homo-XNA duplexes adopt a variety of structures. (D) Representative XNA-only heteroduplexes. FAF:FAF stands for FANA(F)-ANA(A)-FANA(F) XNA:XNA heteroduplex. Alt and chim indicate the alternated or chimeric order of FANA-segments in the duplex sequences respectively. The depicted duplexes have the following PDB ID codes in the Protein Data Bank (http://www.rcsb.org): B-DNA (3BSE); DNA:RNA (1EFS); A-RNA (3ND4); ANA(purple):RNA (2KP3); CeNA(blue):RNA (3KNC); FANA(violet):RNA (2KP4); HNA(yellow):RNA (2BJ6); LNA(cyan):RNA (1H0Q); PNA(orange):DNA (1PDT); PNA(orange):RNA (176D); CeNA:CeNA (blue, 2H0N); hDNA:hDNA (sky blue, 2H9S); FRNA:FRNA (magenta, 3P4A); GNA:GNA (red, 2XC6); HNA:HNA (yellow, 481D); LNA:LNA (cyan, 2×2Q); PNA:PNA (orange, 2K4G), TNA:TNA (green, coordinates not deposited in the PDB [...]); dXyNA:dXyNA (brown, coordinates not deposited in the PDB [...]); XyNA:XyNA (light green, 2N4J); FAF:FAF (FANA in violet, ANA in purple, 2LSC), FRNA:FANA (alt) (FRNA in magenta, FANA in violet, 2M8A); FRNA:FANA (chim) (FRNA in magenta, FANA in violet, 2M84)."[6]
Research
Hypothesis:
- As both ribose and deoxyribose nucleic acids exist, each pentose or hexose sugar should be usable to make a nucleic acid.
Control groups

The findings demonstrate a statistically systematic change from the status quo or the control group.
“In the design of experiments, treatments [or special properties or characteristics] are applied to [or observed in] experimental units in the treatment group(s).[11] In comparative experiments, members of the complementary group, the control group, receive either no treatment or a standard treatment.[12]"[13]
Proof of concept
Def. a “short and/or incomplete realization of a certain method or idea to demonstrate its feasibility"[14] is called a proof of concept.
Def. evidence that demonstrates that a concept is possible is called proof of concept.
The proof-of-concept structure consists of
- background,
- procedures,
- findings, and
- interpretation.[15]
See also
References
- ↑ "phosphoric acid, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. November 10, 2012. Retrieved 2013-04-19.
- 1 2 3 4 "Phosphoric acid, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 9, 2013. Retrieved 2013-04-19.
- ↑ "polyphosphate, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. August 30, 2012. Retrieved 2013-04-19.
- ↑ "deoxyribose, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. April 1, 2013. Retrieved 2013-04-19.
- ↑ "Nucleobase, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. February 22, 2013. Retrieved 2013-04-20.
- 1 2 3 Irina Anosova, Ewa A. Kowal, Matthew R. Dunn, John C. Chaput, Wade D. Van Horn1, and Martin Egli (15 December 2015). "The structural diversity of artificial genetic polymers". Nucleic Acids Research. doi:10.1093/nar/gkv1472. http://nar.oxfordjournals.org/content/early/2015/12/15/nar.gkv1472.full. Retrieved 2016-01-21.
- ↑ "nucleic acid, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. January 12, 2013. Retrieved 2013-04-19.
- ↑ "threose nucleic acid, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. November 14, 2012. Retrieved 2013-04-19.
- ↑ Mayumi Kataoka, Yasuo Kouda, Kousuke Sato, Noriaki Minakawaa and Akira Matsuda (14 August 2011). "Highly efficient enzymatic synthesis of 3′-deoxyapionucleic acid (apioNA) having the four natural nucleobases". Chemical Communications 47 (30): 8700-2. doi:10.1039/C1CC12980E. http://pubs.rsc.org/en/content/articlelanding/2011/cc/c1cc12980e#!divAbstract. Retrieved 2016-01-19.
- 1 2 3 Mohitosh Maiti, Munmun Maiti, Christine Knies, Shrinivas Dumbre, Eveline Lescrinier, Helmut Rosemeyer, Arnout Ceulemans and Piet Herdewijn (13 July 2015). "Xylonucleic acid: synthesis, structure, and orthogonal pairing properties". Nucleic Acids Research 43: 7189-200. doi:10.1093/nar/gkv719. https://nar.oxfordjournals.org/content/early/2015/07/13/nar.gkv719.full. Retrieved 2016-01-21.
- ↑ Klaus Hinkelmann, Oscar Kempthorne (2008). Design and Analysis of Experiments, Volume I: Introduction to Experimental Design (2nd ed.). Wiley. ISBN 978-0-471-72756-9. http://books.google.com/?id=T3wWj2kVYZgC&printsec=frontcover.
- ↑ R. A. Bailey (2008). Design of comparative experiments. Cambridge University Press. ISBN 978-0-521-68357-9. http://www.cambridge.org/uk/catalogue/catalogue.asp?isbn=9780521683579.
- ↑ "Treatment and control groups, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 18, 2012. Retrieved 2012-05-31.
- ↑ "proof of concept, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. November 10, 2012. Retrieved 2013-01-13.
- ↑ Ginger Lehrman and Ian B Hogue, Sarah Palmer, Cheryl Jennings, Celsa A Spina, Ann Wiegand, Alan L Landay, Robert W Coombs, Douglas D Richman, John W Mellors, John M Coffin, Ronald J Bosch, David M Margolis (August 13, 2005). "Depletion of latent HIV-1 infection in vivo: a proof-of-concept study". Lancet 366 (9485): 549-55. doi:10.1016/S0140-6736(05)67098-5. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1894952/. Retrieved 2012-05-09.
External links
- African Journals Online
- Bing Advanced search
- GenomeNet KEGG database
- Google Books
- Google scholar Advanced Scholar Search
- Home - Gene - NCBI
- JSTOR
- Lycos search
- NCBI All Databases Search
- NCBI Site Search
- Office of Scientific & Technical Information
- PsycNET
- PubChem Public Chemical Database
- Questia - The Online Library of Books and Journals
- SAGE journals online
- Scirus for scientific information only advanced search
- SpringerLink
- Taylor & Francis Online
- WikiDoc The Living Textbook of Medicine
- Wiley Online Library Advanced Search
- Yahoo Advanced Web Search
| |||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||
![]() This is a research project at http://en.wikiversity.org
This is a research project at http://en.wikiversity.org
| |
Development status: this resource is experimental in nature. |
| |
Educational level: this is a research resource. |
| |
Resource type: this resource is an article. |
| |
Resource type: this resource contains a lecture or lecture notes. |
| |
Subject classification: this is a biochemistry resource. |
| |
Subject classification: this is a genetics resource. |
| |
Subject classification: this is a medicine resource. |