Gene transcriptions/Boxes
< Gene transcriptions"[A] repeating sequence of nucleotides that forms a transcription or a regulatory signal"[1] is a box.
Genetics

Genetics involves the expression, transmission, and variation of inherited characteristics.
Def. a "branch of biology that deals with the transmission and variation of inherited characteristics, in particular chromosomes and DNA"[2] is called genetics.
AGC boxes

"[T]he AGC box (10), GCC element (11), or AGCCGCC sequence (13), is an ethylene-responsive element found in the promoters of a large number of [pathogenesis related] PR genes".[3]
ATA boxes
The ATA box is a variant of the TATA box that appears in the globin and other genes. Instead of a sequence TATA as in the TATA box, the ATA box lacks the first thymine (T) and may be tissue specific.
An ATA box may have the sequence AAATAT.[4]
CAAT boxes

"[A] CCAAT box (also sometimes abbreviated a CAAT box or CAT box) is a distinct pattern of nucleotides"[5] along the template strand of DNA in eukaryotes.
CArG boxes
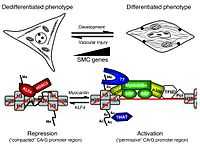
CArG boxes are present in the promoters of smooth muscle cell genes.
C/D boxes
"Located within the introns of very long transcripts extending downstream of SNRPN, there are clusters of paternally expressed C/D box–containing snoRNAs that are highly expressed in the brain5,6."[6]
CENP-B boxes
CGCG boxes
Chromoboxes
Notation: let chromo stand for chromatin organization modifier.[7]
A "[c]hromatin organization modifier (chromo) domain is a conserved region of around 50 amino acids found in a variety of chromosomal proteins, which appear to play a role in the functional organization of the eukaryotic nucleus."[8]
Enhancer boxes

"An E-box (Enhancer Box) is a DNA sequence which usually lies upstream of a gene in a promoter region."[9]
E2 boxes
F boxes
Forkhead boxes
Forkhead "is named for the Drosophila fork head protein, a transcription factor which promotes terminal rather than segmental development."[10]
Fur boxes
G boxes
GC boxes
"[A] GC box is a distinct pattern of nucleotides found in the promoter region of some eukaryotic genes upstream of the TATA box and approximately 110 bases upstream from the transcription initiation site."[11]
H boxes
An H box has a consensus sequence of 3'-ACACCA-5'.[12]
HMG boxes
HNG boxes
HY boxes
A core responsive element is the hypertrophy region HY box between -89 and -60 nucleotides (nts) upstream from the transcription start site.[13]
MADS boxes
"The MADS-box encodes a novel type of DNA-binding domain found so far in a diverse group of transcription factors from yeast, animals, and seed plants."[14]
"The MADS-box comprises 180 nucleotides, encoding 60 amino acids [...] MADS is an acronym for the four DNA-binding proteins MCM1 [minichromosome maintenance gene 1], AGAMOUS [...], DEFICIENS [...], and SRF [serum response factor]."[14]
The "[Antirrhinum majus mutant squamosa(squa)] SQUA is a member of a family of transcription factors which contain the MADS-box, a conserved DNA binding domain."[15]
The "MADS-box is [...] AGAGGGAAAGTACAACTGAAGAGGATAGAGAACAAGATCAATAGACAGGTGACTTT CTCAAAGAGGAGAGGTGGATTGTTGAAAAAAGCTCATGAGCTCTCTGTGCTTTGTG ATGCTGAAGTGGCTCTTATTGTCTTCTCTAATAAGGGGAAGCTATTTGAGTATTCT ACTGAT",[15] which has 174 nucleotides (nts) and begins with the nucleotides for the amino acids RGK.[15] The six nucleotides following the MADS box are "TCTTGC"[15] which may be the additional six needed to get to 180 nts.
P boxes
Pribnow boxes
TACTAAC boxes
T boxes
The T box is a DNA-binding domain.[16]
"T-box genes encode transcription factors involved in the regulation of developmental processes."[16]
TATA boxes

"The TATA box (also called Goldberg-Hogness box)[17] is a DNA sequence (cis-regulatory element) found in the promoter region of genes in archaea and eukaryotes;[18] approximately 24% of human genes contain a TATA box within the core promoter.[19]"[20].
The TATA box is a "binding site of either general transcription factors or histones"[20].
U boxes
"The U box is a domain of ∼70 amino acids that is present in proteins from yeast to humans."[21]
"The prototype U box protein, yeast Ufd2, was identified as a ubiquitin chain assembly factor that cooperates with a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-protein ligase (E3) to catalyze ubiquitin chain formation on artificial substrates."[21]
"The UFD2 protein and its homologs in other eukaryotes share a conserved domain designated the ‘U box’."[22]
"The U box mediates the interaction of UFD2 with ubiquitin conjugated proteins [...] the U box is a derived version of the RING-finger domain that lacks the hallmark metal-chelating residues of the latter [5,6] but is likely to function similarly to the RING-finger in mediating ubiquitin-conjugation of protein substrates."[22]
X boxes
Y boxes
Research
Hypothesis:
- A1BG is not transcribed by any of the boxes.
Control groups

The findings demonstrate a statistically systematic change from the status quo or the control group.
“In the design of experiments, treatments [or special properties or characteristics] are applied to [or observed in] experimental units in the treatment group(s).[23] In comparative experiments, members of the complementary group, the control group, receive either no treatment or a standard treatment.[24]"[25]
Proof of concept
Def. a “short and/or incomplete realization of a certain method or idea to demonstrate its feasibility"[26] is called a proof of concept.
Def. evidence that demonstrates that a concept is possible is called proof of concept.
The proof-of-concept structure consists of
- background,
- procedures,
- findings, and
- interpretation.[27]
See also
References
- ↑ "Box (disambiguation), In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 23, 2013. Retrieved 2013-06-15.
- ↑ "genetics, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. April 16. 2014. Retrieved 2014-05-07.
- ↑ Michael Büttner and Karam B. Singh (May 27, 1997). "Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein". Proceedings of the National Academy of Sciences of the United States of America 94 (11): 5961-6. http://www.pnas.org/content/94/11/5961.long. Retrieved 2014-05-02.
- ↑ Shigemi Kimura, Kuniya Abe, Misao Suzuki, Masakatsu Ogawa, Kowashi Yoshioka, Tadasi Kaname, Teruhisa Miike and Ken-ichi Yamamura (June 1997). "A 900 bp genomic region from the mouse dystrophin promoter directs lacZ reporter expression only to the right heart of transgenic mice". Development, Growth & Differentiation 39 (3): 257-65. doi:10.1046/j.1440-169X.1997.t01-2-00001.x. http://onlinelibrary.wiley.com/doi/10.1046/j.1440-169X.1997.t01-2-00001.x/full. Retrieved 2013-06-28.
- ↑ "CAAT box, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 8, 2013. Retrieved 2013-04-14.
- ↑ Trilochan Sahoo, Daniela del Gaudio, Jennifer R German, Marwan Shinawi, Sarika U Peters, Richard E Person, Adolfo Garnica, Sau Wai Cheung, and Arthur L Beaudet (June 2008). "Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster". Nat. Genet. 40 (6): 719-21. doi:10.1038/ng.158. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2705197/. Retrieved 2014-06-08.
- ↑ Jonathan J. H. Pearce, Prim B. Singh and Stephen J. Gaunt (April 1, 1992). "The mouse has a Polycomb-like chromobox gene". Development 114 (4): 921-9. PMID 1352241. http://dev.biologists.org/content/114/4/921.full.pdf. Retrieved 2013-06-15.
- ↑ NCBI (June 9, 2013). "CBX5 chromobox homolog 5 [ Homo sapiens (human) ]". U.S. National Library of Medicine, 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information. Retrieved 2013-06-15.
- ↑ "E-box, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 13, 2013. Retrieved 2013-04-17.
- ↑ NCBI (June 9, 2013). "FOXP3 forkhead box P3 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2013-06-15.
- ↑ "GC box, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. June 23, 2012. Retrieved 2013-01-27.
- ↑ Timofey S. Rozhdestvensky, Thean Hock Tang, Inna V. Tchirkova, Jürgen Brosius, Jean‐Pierre Bachellerie and Alexander Hüttenhofer (2003). "Binding of L7Ae protein to the K‐turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea". Nucleic Acids Research 31 (3): 869-77. doi:10.1093/nar/gkg175. http://nar.oxfordjournals.org/content/31/3/869.long. Retrieved 2014-06-08.
- ↑ Akiro Higashikawa, Taku Saito, Toshiyuki Ikeda, Satoru Kamekura, Naohiro Kawamura, Akinori Kan, Yasushi Oshima, Shinsuke Ohba, Naoshi Ogata, Katsushi Takeshita, Kozo Nakamura, Ung-Il Chung, Hiroshi Kawaguchi (January 2009). "Identification of the core element responsive to runt-related transcription factor 2 in the promoter of human type x collagen gene". Arthritis & Rheumatism 60 (1): 166-78. doi:10.1002/art.24243. PMID 19116917. http://onlinelibrary.wiley.com/doi/10.1002/art.24243/full. Retrieved 2013-06-18.
- 1 2 Günter Theißen, Jan T. Kim, Heinz Saedler (1 November 1996). "Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes". Journal of Molecular Evolution 43 (5): 484-516. doi:10.1007/BF02337521. http://link.springer.com/article/10.1007/BF02337521. Retrieved 2015-03-31.
- 1 2 3 4 Peter Huijser, Joachim Klein, Wolf-Ekkehard Lonnig, Hans Meijer, Heinz Saedler and Hans Sommer (April 1992). "Bracteomania,an inflorescence anomaly,is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus". The EMBO Journal 11 (4): 1239-49. PMID 556572. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC556572/pdf/emboj00089-0025.pdf. Retrieved 2015-04-01.
- 1 2 NCBI (June 9, 2013). "TBX2 T-box 2 [ Homo sapiens (human) ]". 8600 Rockville Pike, Bethesda MD, 20894 USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 2013-06-15.
- ↑ Lifton RP, Goldberg ML, Karp RW, Hogness DS (1978). "The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications". Cold Spring Harb Symp Quant Biol 42: 1047–51. PMID 98262.
- ↑ Stephen T. Smale and James T. Kadonaga (July 2003). "The RNA Polymerase II Core Promoter". Annual Review of Biochemistry 72 (1): 449-79. doi:10.1146/annurev.biochem.72.121801.161520. PMID 12651739. http://www.annualreviews.org/doi/abs/10.1146/annurev.biochem.72.121801.161520. Retrieved 2012-05-07.
- ↑ C Yang, E Bolotin, T Jiang, FM Sladek, E Martinez (March 2007). "Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters". Gene 389 (1): 52–65. doi:10.1016/j.gene.2006.09.029. PMID 17123746. PMC 1955227. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1955227/?tool=pubmed.
- 1 2 "TATA box, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. April 30, 2012. Retrieved 2012-05-10.
- 1 2 Shigetsugu Hatakeyama, Masayoshi Yada, Masaki Matsumoto, Noriko Ishida and Kei-Ichi Nakayama (August 31, 2001). "U Box Proteins as a New Family of Ubiquitin-Protein Ligases". The Journal of Biological Chemistry 276: 33111-20. doi:10.1074/jbc.M102755200. http://www.jbc.org/content/276/35/33111.full. Retrieved 2014-06-16.
- 1 2 L. Aravind and Eugene V. Koonin (2000). "The U box is a modified RING finger — a common domain in ubiquitination". Current Biology 10 (4): R132-4.
- ↑ Klaus Hinkelmann, Oscar Kempthorne (2008). Design and Analysis of Experiments, Volume I: Introduction to Experimental Design (2nd ed.). Wiley. ISBN 978-0-471-72756-9. http://books.google.com/?id=T3wWj2kVYZgC&printsec=frontcover.
- ↑ R. A. Bailey (2008). Design of comparative experiments. Cambridge University Press. ISBN 978-0-521-68357-9. http://www.cambridge.org/uk/catalogue/catalogue.asp?isbn=9780521683579.
- ↑ "Treatment and control groups, In: Wikipedia". San Francisco, California: Wikimedia Foundation, Inc. May 18, 2012. Retrieved 2012-05-31.
- ↑ "proof of concept, In: Wiktionary". San Francisco, California: Wikimedia Foundation, Inc. November 10, 2012. Retrieved 2013-01-13.
- ↑ Ginger Lehrman and Ian B Hogue, Sarah Palmer, Cheryl Jennings, Celsa A Spina, Ann Wiegand, Alan L Landay, Robert W Coombs, Douglas D Richman, John W Mellors, John M Coffin, Ronald J Bosch, David M Margolis (August 13, 2005). "Depletion of latent HIV-1 infection in vivo: a proof-of-concept study". Lancet 366 (9485): 549-55. doi:10.1016/S0140-6736(05)67098-5. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1894952/. Retrieved 2012-05-09.
External links
- African Journals Online
- Bing Advanced search
- GenomeNet KEGG database
- Google Books
- Google scholar Advanced Scholar Search
- Home - Gene - NCBI
- JSTOR
- Lycos search
- NCBI All Databases Search
- NCBI Site Search
- Office of Scientific & Technical Information
- PubChem Public Chemical Database
- Questia - The Online Library of Books and Journals
- SAGE journals online
- Scirus for scientific information only advanced search
- SpringerLink
- Taylor & Francis Online
- WikiDoc The Living Textbook of Medicine
- Wiley Online Library Advanced Search
- Yahoo Advanced Web Search